Parvovirus diversity and DNA damage responses
- PMID: 23293137
- PMCID: PMC3552509
- DOI: 10.1101/cshperspect.a012989
Parvovirus diversity and DNA damage responses
Abstract
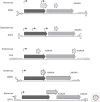
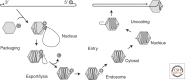
Parvoviruses have a linear single-stranded DNA genome, around 5 kb in length, with short imperfect terminal palindromes that fold back on themselves to form duplex hairpin telomeres. These contain most of the cis-acting information required for viral "rolling hairpin" DNA replication, an evolutionary adaptation of rolling-circle synthesis in which the hairpins create duplex replication origins, prime complementary strand synthesis, and act as hinges to reverse the direction of the unidirectional cellular fork. Genomes are packaged vectorially into small, rugged protein capsids ~260 Å in diameter, which mediate their delivery directly into the cell nucleus, where they await their host cell's entry into S phase under its own cell cycle control. Here we focus on genus-specific variations in genome structure and replication, and review host cell responses that modulate the nuclear environment.
Figures
References
-
- Cervelli T, Palacios JA, Zentilin L, Mano M, Schwartz RA, Weitzman MD, Giacca M 2008. Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins. J Cell Science 121: 349–357 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
