Major chromosomal breakpoint intervals in breast cancer co-localize with differentially methylated regions
- PMID: 23293768
- PMCID: PMC3530719
- DOI: 10.3389/fonc.2012.00197
Major chromosomal breakpoint intervals in breast cancer co-localize with differentially methylated regions
Abstract
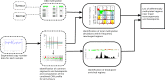
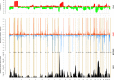
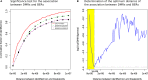
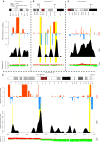
Solid tumors exhibit chromosomal rearrangements resulting in gain or loss of multiple chromosomal loci (copy number variation, or CNV), and translocations that occasionally result in the creation of novel chimeric genes. In the case of breast cancer, although most individual tumors each have unique CNV landscape, the breakpoints, as measured over large datasets, appear to be non-randomly distributed in the genome. Breakpoints show a significant regional concentration at genomic loci spanning perhaps several megabases. The proximal cause of these breakpoint concentrations is a subject of speculation, but is, as yet, largely unknown. To shed light on this issue, we have performed a bio-statistical analysis on our previously published data for a set of 119 breast tumors and normal controls (Wiedswang et al., 2003), where each sample has both high-resolution CNV and methylation data. The method examined the distribution of closeness of breakpoint regions with differentially methylated regions (DMR), coupled with additional genomic parameters, such as repeat elements and designated "fragile sites" in the reference genome. Through this analysis, we have identified a set of 93 regional loci called breakpoint enriched DMR (BEDMRs) characterized by altered DNA methylation in cancer compared to normal cells that are associated with frequent breakpoint concentrations within a distance of 1 Mb. BEDMR loci are further associated with local hypomethylation (66%), concentrations of the Alu SINE repeats within 3 Mb (35% of the cases), and tend to occur near a number of cancer related genes such as the protocadherins, AKT1, DUB3, GAB2. Furthermore, BEDMRs seem to deregulate members of the histone gene family and chromatin remodeling factors, e.g., JMJD1B, which might affect the chromatin structure and disrupt coordinate signaling and repair. From this analysis we propose that preference for chromosomal breakpoints is related to genome structure coupled with alterations in DNA methylation and hence, chromatin structure, associated with tumorigenesis.
Keywords: Alu repeat element; DNA methylation; breast cancer; copy number variation; genome instability; multi-modal analysis.
Figures
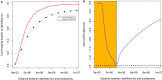
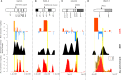
References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

