Structure of Pisum sativum Rubisco with bound ribulose 1,5-bisphosphate
- PMID: 23295478
- PMCID: PMC3539695
- DOI: 10.1107/S1744309112047549
Structure of Pisum sativum Rubisco with bound ribulose 1,5-bisphosphate
Abstract
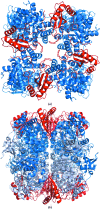
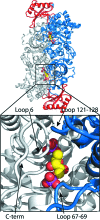
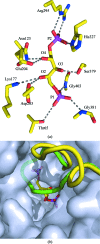
The first structure of a ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from a pulse crop is reported. Rubisco was purified from Pisum sativum (garden pea) and diffraction-quality crystals were obtained by hanging-drop vapour diffusion in the presence of the substrate ribulose 1,5-bisphosphate. X-ray diffraction data were recorded to 2.20 Å resolution from a single crystal at the Canadian Light Source. The overall quaternary structure of non-activated P. sativum Rubisco highlights the conservation of the form I Rubisco hexadecameric complex. The electron density places the substrate in the active site at the interface of the large-subunit dimers. Lys201 in the active site is not carbamylated as expected for this non-activated structure. Some heterogeneity in the small-subunit sequence is noted, as well as possible variations in the conformation and contacts of ribulose 1,5-bisphosphate in the large-subunit active sites. Overall, the active-site conformation most closely correlates with the `closed' conformation observed in other substrate/inhibitor-bound Rubisco structures.
Figures

Similar articles
-
A unique structural domain in Methanococcoides burtonii ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) acts as a small subunit mimic.J Biol Chem. 2017 Apr 21;292(16):6838-6850. doi: 10.1074/jbc.M116.767145. Epub 2017 Jan 30. J Biol Chem. 2017. PMID: 28154188 Free PMC article.
-
The structure of the complex between rubisco and its natural substrate ribulose 1,5-bisphosphate.J Mol Biol. 1997 Jan 31;265(4):432-44. doi: 10.1006/jmbi.1996.0738. J Mol Biol. 1997. PMID: 9034362
-
Subunit interface dynamics in hexadecameric rubisco.J Mol Biol. 2011 Sep 2;411(5):1083-98. doi: 10.1016/j.jmb.2011.06.052. Epub 2011 Jul 6. J Mol Biol. 2011. PMID: 21745478
-
Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase.Arch Biochem Biophys. 2003 Jun 15;414(2):141-9. doi: 10.1016/s0003-9861(03)00171-1. Arch Biochem Biophys. 2003. PMID: 12781765 Review.
-
Structure and function of Rubisco.Plant Physiol Biochem. 2008 Mar;46(3):275-91. doi: 10.1016/j.plaphy.2008.01.001. Epub 2008 Jan 12. Plant Physiol Biochem. 2008. PMID: 18294858 Review.
Cited by
-
The small subunit of Rubisco and its potential as an engineering target.J Exp Bot. 2023 Jan 11;74(2):543-561. doi: 10.1093/jxb/erac309. J Exp Bot. 2023. PMID: 35849331 Free PMC article. Review.
-
Application of an improved proteomics method for abundant protein cleanup: molecular and genomic mechanisms study in plant defense.Mol Cell Proteomics. 2013 Nov;12(11):3431-42. doi: 10.1074/mcp.M112.025213. Epub 2013 Aug 13. Mol Cell Proteomics. 2013. PMID: 23943779 Free PMC article.
-
Identification of Interactions between Abscisic Acid and Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase.PLoS One. 2015 Jul 21;10(7):e0133033. doi: 10.1371/journal.pone.0133033. eCollection 2015. PLoS One. 2015. PMID: 26197050 Free PMC article.
-
Structure of Rubisco from Arabidopsis thaliana in complex with 2-carboxyarabinitol-1,5-bisphosphate.Acta Crystallogr D Struct Biol. 2018 Jan 1;74(Pt 1):1-9. doi: 10.1107/S2059798317017132. Epub 2018 Jan 1. Acta Crystallogr D Struct Biol. 2018. PMID: 29372894 Free PMC article.
References
-
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221. - PubMed
-
- Andersson, I. (1996). J. Mol. Biol. 259, 160–174. - PubMed
-
- Andersson, I. (2008). J. Exp. Bot. 59, 1555–1568. - PubMed
-
- Andersson, I. & Backlund, A. (2008). Plant Physiol. Biochem. 46, 275–291. - PubMed
-
- Andersson, I., Knight, S., Schneider, G., Lindqvist, Y., Lundqvist, T., Brändén, C.-I. & Lorimer, G. H. (1989). Nature (London), 337, 231–234.
Publication types
MeSH terms
Substances
Associated data
- Actions

