Structural and functional evidence that Rad4 competes with Rad2 for binding to the Tfb1 subunit of TFIIH in NER
- PMID: 23295669
- PMCID: PMC3575800
- DOI: 10.1093/nar/gks1321
Structural and functional evidence that Rad4 competes with Rad2 for binding to the Tfb1 subunit of TFIIH in NER
Abstract
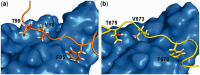
XPC/Rad4 (human/yeast) recruits transcription faction IIH (TFIIH) to the nucleotide excision repair (NER) complex through interactions with its p62/Tfb1 and XPB/Ssl2 subunits. TFIIH then recruits XPG/Rad2 through interactions with similar subunits and the two repair factors appear to be mutually exclusive within the NER complex. Here, we show that Rad4 binds the PH domain of the Tfb1 (Tfb1PH) with high affinity. Structural characterization of a Rad4-Tfb1PH complex demonstrates that the Rad4-binding interface is formed using a motif similar to one used by Rad2 to bind Tfb1PH. In vivo studies in yeast demonstrate that the N-terminal Tfb1-binding motif and C-terminal TFIIH-binding motif of Rad4 are both crucial for survival following exposure to UV irradiation. Together, these results support the hypothesis that XPG/Rad2 displaces XPC/Rad4 from the repair complex in part through interactions with the Tfb1/p62 subunit of TFIIH. The Rad4-Tfb1PH structure also provides detailed information regarding, not only the interplay of TFIIH recruitment to the NER, but also links the role of TFIIH in NER and transcription.
Figures



Similar articles
-
Structural and functional characterization of interactions involving the Tfb1 subunit of TFIIH and the NER factor Rad2.Nucleic Acids Res. 2012 Jul;40(12):5739-50. doi: 10.1093/nar/gks194. Epub 2012 Feb 28. Nucleic Acids Res. 2012. PMID: 22373916 Free PMC article.
-
Interplay of the Tfb1 pleckstrin homology domain with Rad2 and Rad4 in transcription coupled and global genomic nucleotide excision repair.Nucleic Acids Res. 2024 Jun 24;52(11):6333-6346. doi: 10.1093/nar/gkae286. Nucleic Acids Res. 2024. PMID: 38634797 Free PMC article.
-
NMR structure of the amino-terminal domain from the Tfb1 subunit of TFIIH and characterization of its phosphoinositide and VP16 binding sites.Biochemistry. 2005 May 31;44(21):7678-86. doi: 10.1021/bi050099s. Biochemistry. 2005. PMID: 15909982
-
Molecular basis for damage recognition and verification by XPC-RAD23B and TFIIH in nucleotide excision repair.DNA Repair (Amst). 2018 Nov;71:33-42. doi: 10.1016/j.dnarep.2018.08.005. Epub 2018 Aug 23. DNA Repair (Amst). 2018. PMID: 30174301 Free PMC article. Review.
-
Nucleotide excision repair in yeast.Mutat Res. 2000 Jun 30;451(1-2):13-24. doi: 10.1016/s0027-5107(00)00037-3. Mutat Res. 2000. PMID: 10915862 Review.
Cited by
-
Common TFIIH recruitment mechanism in global genome and transcription-coupled repair subpathways.Nucleic Acids Res. 2017 Dec 15;45(22):13043-13055. doi: 10.1093/nar/gkx970. Nucleic Acids Res. 2017. PMID: 29069470 Free PMC article.
-
Structural polymorphism of the PH domain in TFIIH.Biosci Rep. 2023 Jul 26;43(7):BSR20230846. doi: 10.1042/BSR20230846. Biosci Rep. 2023. PMID: 37340985 Free PMC article.
-
ETV4 and AP1 Transcription Factors Form Multivalent Interactions with three Sites on the MED25 Activator-Interacting Domain.J Mol Biol. 2017 Oct 13;429(20):2975-2995. doi: 10.1016/j.jmb.2017.06.024. Epub 2017 Jul 17. J Mol Biol. 2017. PMID: 28728983 Free PMC article.
-
Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.Nat Commun. 2021 Jun 7;12(1):3338. doi: 10.1038/s41467-021-23684-x. Nat Commun. 2021. PMID: 34099686 Free PMC article.
-
Structural basis of transcription initiation by RNA polymerase II.Nat Rev Mol Cell Biol. 2015 Mar;16(3):129-43. doi: 10.1038/nrm3952. Epub 2015 Feb 18. Nat Rev Mol Cell Biol. 2015. PMID: 25693126 Review.
References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz T, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2005.
-
- Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. - PubMed
-
- Cartault F, Nava C, Malbrunot AC, Munier P, Hebert JC, N'Guyen P, Djeridi N, Pariaud P, Pariaud J, Dupuy A, et al. A new XPC gene splicing mutation has lead to the highest worldwide prevalence of xeroderma pigmentosum in black Mahori patients. DNA Repair. 2011;10:577–585. - PubMed
-
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. - PubMed
-
- Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

