Human rhinoviruses
- PMID: 23297263
- PMCID: PMC3553670
- DOI: 10.1128/CMR.00077-12
Human rhinoviruses
Abstract
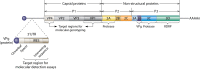
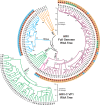
Human rhinoviruses (HRVs), first discovered in the 1950s, are responsible for more than one-half of cold-like illnesses and cost billions of dollars annually in medical visits and missed days of work. Advances in molecular methods have enhanced our understanding of the genomic structure of HRV and have led to the characterization of three genetically distinct HRV groups, designated groups A, B, and C, within the genus Enterovirus and the family Picornaviridae. HRVs are traditionally associated with upper respiratory tract infection, otitis media, and sinusitis. In recent years, the increasing implementation of PCR assays for respiratory virus detection in clinical laboratories has facilitated the recognition of HRV as a lower respiratory tract pathogen, particularly in patients with asthma, infants, elderly patients, and immunocompromised hosts. Cultured isolates of HRV remain important for studies of viral characteristics and disease pathogenesis. Indeed, whether the clinical manifestations of HRV are related directly to viral pathogenicity or secondary to the host immune response is the subject of ongoing research. There are currently no approved antiviral therapies for HRVs, and treatment remains primarily supportive. This review provides a comprehensive, up-to-date assessment of the basic virology, pathogenesis, clinical epidemiology, and laboratory features of and treatment and prevention strategies for HRVs.
Figures







References
-
- Nichol KL, D'Heilly S, Ehlinger E. 2005. Colds and influenza-like illnesses in university students: impact on health, academic and work performance, and health care use. Clin. Infect. Dis. 40:1263–1270 - PubMed
-
- Roelen CAM, Koopmans PC, Notenbomer A, Groothoff JW. 2011. Job satisfaction and short sickness absence due to the common cold. Work 39:305–313 - PubMed
-
- Bertino JS. 2002. Cost burden of viral respiratory infections: issues for formulary decision makers. Am. J. Med. 112:42S–49S doi:10.1016/S0002-9343(01)01063-4 - DOI - PubMed
-
- Fendrick AM, Monto AS, Nightengale B, Sarnes M. 2003. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch. Intern. Med. 163:487–494 - PubMed
-
- Pappas DE, Hendley JO, Hayden FG, Winther B. 2008. Symptom profile of common colds in school-aged children. Pediatr. Infect. Dis. J. 27:8–11 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

