Molecular architecture of a complex between an adhesion protein from the malaria parasite and intracellular adhesion molecule 1
- PMID: 23297413
- PMCID: PMC3581401
- DOI: 10.1074/jbc.M112.416347
Molecular architecture of a complex between an adhesion protein from the malaria parasite and intracellular adhesion molecule 1
Abstract
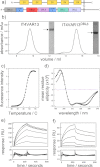
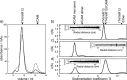
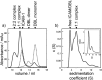
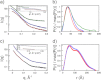
The adhesion of Plasmodium falciparum-infected erythrocytes to human tissues or endothelium is central to the pathology caused by the parasite during malaria. It contributes to the avoidance of parasite clearance by the spleen and to the specific pathologies of cerebral and placental malaria. The PfEMP1 family of adhesive proteins is responsible for this sequestration by mediating interactions with diverse human ligands. In addition, as the primary targets of acquired, protective immunity, the PfEMP1s are potential vaccine candidates. PfEMP1s contain large extracellular ectodomains made from CIDR (cysteine-rich interdomain regions) and DBL (Duffy-binding-like) domains and show extensive variation in sequence, size, and domain organization. Here we use biophysical methods to characterize the entire ∼300-kDa ectodomain from IT4VAR13, a protein that interacts with the host receptor, intercellular adhesion molecule-1 (ICAM-1). We show through small angle x-ray scattering that IT4VAR13 is rigid, elongated, and monomeric. We also show that it interacts with ICAM-1 through the DBLβ domain alone, forming a 1:1 complex. These studies provide a first low resolution structural view of a PfEMP1 ectodomain in complex with its ligand. They show that it combines a modular domain arrangement consisting of individual ligand binding domains, with a defined higher order architecture that exposes the ICAM-1 binding surface to allow adhesion.
Figures

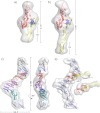
References
-
- WHO (2011) World Malaria Report 2011, Geneva, Switzerland
-
- Turner G. D., Morrison H., Jones M., Davis T. M., Looareesuwan S., Buley I. D., Gatter K. C., Newbold C. I., Pukritayakamee S., Nagachinta B. (1994) An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am. J. Pathol. 145, 1057–1069 - PMC - PubMed
-
- Pongponratn E., Turner G. D., Day N. P., Phu N. H., Simpson J. A., Stepniewska K., Mai N. T., Viriyavejakul P., Looareesuwan S., Hien T. T., Ferguson D. J., White N. J. (2003) An ultrastructural study of the brain in fatal Plasmodium falciparum malaria. Am. J. Trop. Med. Hyg. 69, 345–359 - PubMed
-
- Smith J. D., Chitnis C. E., Craig A. G., Roberts D. J., Hudson-Taylor D. E., Peterson D. S., Pinches R., Newbold C. I., Miller L. H. (1995) Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82, 101–110 - PMC - PubMed
-
- Su X. Z., Heatwole V. M., Wertheimer S. P., Guinet F., Herrfeldt J. A., Peterson D. S., Ravetch J. A., Wellems T. E. (1995) The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82, 89–100 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

