Connecting protein conformational dynamics with catalytic function as illustrated in dihydrofolate reductase
- PMID: 23297871
- PMCID: PMC3773050
- DOI: 10.1021/bi301559q
Connecting protein conformational dynamics with catalytic function as illustrated in dihydrofolate reductase
Abstract
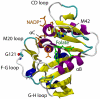
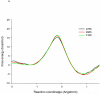
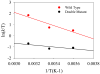
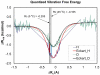
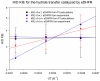
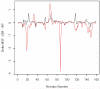
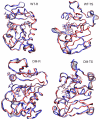
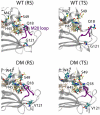
Combined quantum mechanics/molecular mechanics molecular dynamics simulations reveal that the M20 loop conformational dynamics of dihydrofolate reductase (DHFR) is severely restricted at the transition state of the hydride transfer as a result of the M42W/G121V double mutation. Consequently, the double-mutant enzyme has a reduced entropy of activation, i.e., increased entropic barrier, and altered temperature dependence of kinetic isotope effects in comparison with those of wild-type DHFR. Interestingly, in both wild-type DHFR and the double mutant, the average donor-acceptor distances are essentially the same in the Michaelis complex state (~3.5 Å) and the transition state (2.7 Å). It was found that an additional hydrogen bond is formed to stabilize the M20 loop in the closed conformation in the M42W/G121V double mutant. The computational results reflect a similar aim designed to knock out precisely the dynamic flexibility of the M20 loop in a different double mutant, N23PP/S148A.
Figures



References
-
- Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem. Rev. 2006;106:3055–3079. - PubMed
-
- Hay S, Scrutton NS. Good vibrations in enzyme-catalysed reactions. Nat Chem. 2012;4:161–168. - PubMed
-
- Nagel ZD, Klinman JP. A 21(st) century revisionist’s view at a turning point in enzymology. Nature Chem. Biol. 2009;5:543–550. - PubMed
-
- Glowacki DR, Harvey JN, Mulholland AJ. Taking Ockham’s razor to enzyme dynamics and catalysis. Nat Chem. 2012;4:169–176. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

