Deformable registration of the inflated and deflated lung in cone-beam CT-guided thoracic surgery: initial investigation of a combined model- and image-driven approach
- PMID: 23298134
- PMCID: PMC3537709
- DOI: 10.1118/1.4767757
Deformable registration of the inflated and deflated lung in cone-beam CT-guided thoracic surgery: initial investigation of a combined model- and image-driven approach
Abstract
Purpose: Surgical resection is the preferred modality for curative treatment of early stage lung cancer, but localization of small tumors (<10 mm diameter) during surgery presents a major challenge that is likely to increase as more early-stage disease is detected incidentally and in low-dose CT screening. To overcome the difficulty of manual localization (fingers inserted through intercostal ports) and the cost, logistics, and morbidity of preoperative tagging (coil or dye placement under CT-fluoroscopy), the authors propose the use of intraoperative cone-beam CT (CBCT) and deformable image registration to guide targeting of small tumors in video-assisted thoracic surgery (VATS). A novel algorithm is reported for registration of the lung from its inflated state (prior to pleural breach) to the deflated state (during resection) to localize surgical targets and adjacent critical anatomy.
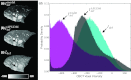
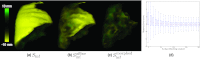
Methods: The registration approach geometrically resolves images of the inflated and deflated lung using a coarse model-driven stage followed by a finer image-driven stage. The model-driven stage uses image features derived from the lung surfaces and airways: triangular surface meshes are morphed to capture bulk motion; concurrently, the airways generate graph structures from which corresponding nodes are identified. Interpolation of the sparse motion fields computed from the bounding surface and interior airways provides a 3D motion field that coarsely registers the lung and initializes the subsequent image-driven stage. The image-driven stage employs an intensity-corrected, symmetric form of the Demons method. The algorithm was validated over 12 datasets, obtained from porcine specimen experiments emulating CBCT-guided VATS. Geometric accuracy was quantified in terms of target registration error (TRE) in anatomical targets throughout the lung, and normalized cross-correlation. Variations of the algorithm were investigated to study the behavior of the model- and image-driven stages by modifying individual algorithmic steps and examining the effect in comparison to the nominal process.
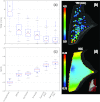
Results: The combined model- and image-driven registration process demonstrated accuracy consistent with the requirements of minimally invasive VATS in both target localization (∼3-5 mm within the target wedge) and critical structure avoidance (∼1-2 mm). The model-driven stage initialized the registration to within a median TRE of 1.9 mm (95% confidence interval (CI) maximum = 5.0 mm), while the subsequent image-driven stage yielded higher accuracy localization with 0.6 mm median TRE (95% CI maximum = 4.1 mm). The variations assessing the individual algorithmic steps elucidated the role of each step and in some cases identified opportunities for further simplification and improvement in computational speed.
Conclusions: The initial studies show the proposed registration method to successfully register CBCT images of the inflated and deflated lung. Accuracy appears sufficient to localize the target and adjacent critical anatomy within ∼1-2 mm and guide localization under conditions in which the target cannot be discerned directly in CBCT (e.g., subtle, nonsolid tumors). The ability to directly localize tumors in the operating room could provide a valuable addition to the VATS arsenal, obviate the cost, logistics, and morbidity of preoperative tagging, and improve patient safety. Future work includes in vivo testing, optimization of workflow, and integration with a CBCT image guidance system.
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

