Ubiquitination by the membrane-associated RING-CH-8 (MARCH-8) ligase controls steady-state cell surface expression of tumor necrosis factor-related apoptosis inducing ligand (TRAIL) receptor 1
- PMID: 23300075
- PMCID: PMC3585101
- DOI: 10.1074/jbc.M112.448209
Ubiquitination by the membrane-associated RING-CH-8 (MARCH-8) ligase controls steady-state cell surface expression of tumor necrosis factor-related apoptosis inducing ligand (TRAIL) receptor 1
Abstract
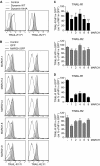
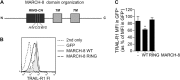
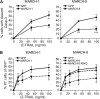
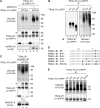
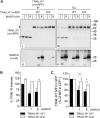
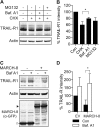
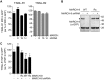
The eleven members of the membrane-associated RING-CH (MARCH) ubiquitin ligase family are relatively unexplored. Upon exogenous (over)expression, a number of these ligases can affect the trafficking of membrane molecules. However, only for MARCH-1 endogenous functions have been demonstrated. For the other endogenous MARCH proteins, no functions or substrates are known. We report here that TRAIL-R1 is a physiological substrate of the endogenous MARCH-8 ligase. Human TRAIL-R1 and R2 play a role in immunosurveillance and are targets for cancer therapy, because they selectively induce apoptosis in tumor cells. We demonstrate that TRAIL-R1 is down-regulated from the cell surface, with great preference over TRAIL-R2, by exogenous expression of MARCH ligases that are implicated in endosomal trafficking, such as MARCH-1 and -8. MARCH-8 attenuated TRAIL-R1 cell surface expression and apoptosis signaling by virtue of its ligase activity. This suggested that ubiquitination of TRAIL-R1 was instrumental in its down-regulation by MARCH-8. Indeed, in cells with endogenous MARCH expression, TRAIL-R1 was ubiquitinated at steady-state, with the conserved membrane-proximal lysine 273 as one of the potential acceptor sites. This residue was also essential for the interaction of TRAIL-R1 with MARCH-1 and MARCH-8 and its down-regulation by these ligases. Gene silencing identified MARCH-8 as the endogenous ligase that ubiquitinates TRAIL-R1 and attenuates its cell surface expression. These findings reveal that endogenous MARCH-8 regulates the steady-state cell surface expression of TRAIL-R1.
Figures

Similar articles
-
Negative control of TRAIL-R1 signaling by transforming growth factor β1 in pancreatic tumor cells involves Smad-dependent down regulation of TRAIL-R1.Cell Signal. 2016 Nov;28(11):1652-62. doi: 10.1016/j.cellsig.2016.07.016. Epub 2016 Aug 1. Cell Signal. 2016. PMID: 27492861
-
The TRAIL-receptor-1: TRAIL-receptor-3 and -4 ratio is a predictor for TRAIL sensitivity of cancer cells.Oncol Rep. 2009 May;21(5):1289-95. doi: 10.3892/or_00000353. Oncol Rep. 2009. PMID: 19360306
-
Regulation of TRAIL-receptor expression by the ubiquitin-proteasome system.Int J Mol Sci. 2014 Oct 14;15(10):18557-73. doi: 10.3390/ijms151018557. Int J Mol Sci. 2014. PMID: 25318057 Free PMC article. Review.
-
Ubiquitin-conjugating enzyme E2 D1 (Ube2D1) mediates lysine-independent ubiquitination of the E3 ubiquitin ligase March-I.J Biol Chem. 2018 Mar 16;293(11):3904-3912. doi: 10.1074/jbc.RA117.001322. Epub 2018 Feb 1. J Biol Chem. 2018. PMID: 29414787 Free PMC article.
-
E3 ubiquitin ligases and deubiquitinases as modulators of TRAIL-mediated extrinsic apoptotic signaling pathway.BMB Rep. 2019 Feb;52(2):119-126. doi: 10.5483/BMBRep.2019.52.2.011. BMB Rep. 2019. PMID: 30638181 Free PMC article. Review.
Cited by
-
The Membrane Associated RING-CH Proteins: A Family of E3 Ligases with Diverse Roles through the Cell.Int Sch Res Notices. 2014 Oct 29;2014:637295. doi: 10.1155/2014/637295. eCollection 2014. Int Sch Res Notices. 2014. PMID: 27419207 Free PMC article. Review.
-
Advances in HIV-1 Assembly.Viruses. 2022 Feb 26;14(3):478. doi: 10.3390/v14030478. Viruses. 2022. PMID: 35336885 Free PMC article. Review.
-
Ubiquitin-dependent sorting in endocytosis.Cold Spring Harb Perspect Biol. 2014 Jan 1;6(1):a016808. doi: 10.1101/cshperspect.a016808. Cold Spring Harb Perspect Biol. 2014. PMID: 24384571 Free PMC article. Review.
-
JWA regulates TRAIL-induced apoptosis via MARCH8-mediated DR4 ubiquitination in cisplatin-resistant gastric cancer cells.Oncogenesis. 2017 Jul 3;6(7):e353. doi: 10.1038/oncsis.2017.57. Oncogenesis. 2017. PMID: 28671676 Free PMC article.
-
Further Characterization of the Antiviral Transmembrane Protein MARCH8.Cells. 2024 Apr 17;13(8):698. doi: 10.3390/cells13080698. Cells. 2024. PMID: 38667313 Free PMC article.
References
-
- Smyth M. J., Cretney E., Takeda K., Wiltrout R. H., Sedger L. M., Kayagaki N., Yagita H., Okumura K. (2001) Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) contributes to interferon γ-dependent natural killer cell protection from tumor metastasis. J. Exp. Med. 193, 661–670 - PMC - PubMed
-
- Takeda K., Hayakawa Y., Smyth M. J., Kayagaki N., Yamaguchi N., Kakuta S., Iwakura Y., Yagita H., Okumura K. (2001) Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 7, 94–100 - PubMed
-
- Kimberley F. C., Screaton G. R. (2004) Following a TRAIL. Update on a ligand and its five receptors. Cell Res. 14, 359–372 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

