Spliceosome mutations exhibit specific associations with epigenetic modifiers and proto-oncogenes mutated in myelodysplastic syndrome
- PMID: 23300180
- PMCID: PMC3696609
- DOI: 10.3324/haematol.2012.075325
Spliceosome mutations exhibit specific associations with epigenetic modifiers and proto-oncogenes mutated in myelodysplastic syndrome
Abstract
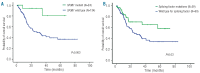
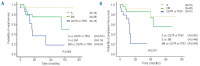
The recent identification of acquired mutations in key components of the spliceosome machinery strongly implicates abnormalities of mRNA splicing in the pathogenesis of myelodysplastic syndromes. However, questions remain as to how these aberrations functionally combine with the growing list of mutations in genes involved in epigenetic modification and cell signaling/transcription regulation identified in these diseases. In this study, amplicon sequencing was used to perform a mutation screen in 154 myelodysplastic syndrome patients using a 22-gene panel, including commonly mutated spliceosome components (SF3B1, SRSF2, U2AF1, ZRSR2), and a further 18 genes known to be mutated in myeloid cancers. Sequencing of the 22-gene panel revealed that 76% (n=117) of the patients had mutations in at least one of the genes, with 38% (n=59) having splicing gene mutations and 49% (n=75) patients harboring more than one gene mutation. Interestingly, single and specific epigenetic modifier mutations tended to coexist with SF3B1 and SRSF2 mutations (P<0.03). Furthermore, mutations in SF3B1 and SRSF2 were mutually exclusive to TP53 mutations both at diagnosis and at the time of disease transformation. Moreover, mutations in FLT3, NRAS, RUNX1, CCBL and C-KIT were more likely to co-occur with splicing factor mutations generally (P<0.02), and SRSF2 mutants in particular (P<0.003) and were significantly associated with disease transformation (P<0.02). SF3B1 and TP53 mutations had varying impacts on overall survival with hazard ratios of 0.2 (P<0.03, 95% CI, 0.1-0.8) and 2.1 (P<0.04, 95% CI, 1.1-4.4), respectively. Moreover, patients with splicing factor mutations alone had a better overall survival than those with epigenetic modifier mutations, or cell signaling/transcription regulator mutations with and without coexisting mutations of splicing factor genes, with worsening prognosis (P<0.001). These findings suggest that splicing factor mutations are maintained throughout disease evolution with emerging oncogenic mutations adversely affecting patients' outcome, implicating spliceosome mutations as founder mutations in myelodysplastic syndromes.
Figures


References
-
- Mufti GJ, Bennett JM, Goasguen J, Bain BJ, Baumann I, Brunning R, et al. Diagnosis and classification of myelodysplastic syndrome: International Working Group on Morphology of myelodysplastic syndrome (IWGM-MDS) consensus proposals for the definition and enumeration of myeloblasts and ring sideroblasts. Haematologica. 2008;93(11):1712–7 - PubMed
-
- Tefferi A, Vardiman JW. Myelodysplastic syndromes. N Engl J Med. 2009;361(19): 1872–85 - PubMed
-
- Corey SJ, Minden MD, Barber DL, Kantarjian H, Wang JC, Schimmer AD. Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat Rev Cancer. 2007;7(2):118–29 - PubMed
-
- Pozdnyakova O, Miron PM, Tang G, Walter O, Raza A, Woda B, Wang SA. Cytogenetic abnormalities in a series of 1,029 patients with primary myelodysplastic syndromes: a report from the US with a focus on some undefined single chromosomal abnormalities. Cancer. 2008;113(12): 3331–40 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

