ELF5 suppresses estrogen sensitivity and underpins the acquisition of antiestrogen resistance in luminal breast cancer
- PMID: 23300383
- PMCID: PMC3531499
- DOI: 10.1371/journal.pbio.1001461
ELF5 suppresses estrogen sensitivity and underpins the acquisition of antiestrogen resistance in luminal breast cancer
Abstract
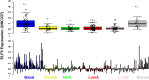
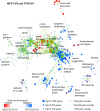
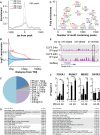
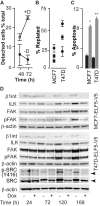
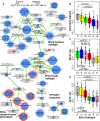
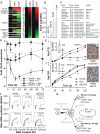
We have previously shown that during pregnancy the E-twenty-six (ETS) transcription factor ELF5 directs the differentiation of mammary progenitor cells toward the estrogen receptor (ER)-negative and milk producing cell lineage, raising the possibility that ELF5 may suppress the estrogen sensitivity of breast cancers. To test this we constructed inducible models of ELF5 expression in ER positive luminal breast cancer cells and interrogated them using transcript profiling and chromatin immunoprecipitation of DNA followed by DNA sequencing (ChIP-Seq). ELF5 suppressed ER and FOXA1 expression and broadly suppressed ER-driven patterns of gene expression including sets of genes distinguishing the luminal molecular subtype. Direct transcriptional targets of ELF5, which included FOXA1, EGFR, and MYC, accurately classified a large cohort of breast cancers into their intrinsic molecular subtypes, predicted ER status with high precision, and defined groups with differential prognosis. Knockdown of ELF5 in basal breast cancer cell lines suppressed basal patterns of gene expression and produced a shift in molecular subtype toward the claudin-low and normal-like groups. Luminal breast cancer cells that acquired resistance to the antiestrogen Tamoxifen showed greatly elevated levels of ELF5 and its transcriptional signature, and became dependent on ELF5 for proliferation, compared to the parental cells. Thus ELF5 provides a key transcriptional determinant of breast cancer molecular subtype by suppression of estrogen sensitivity in luminal breast cancer cells and promotion of basal characteristics in basal breast cancer cells, an action that may be utilised to acquire antiestrogen resistance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. (2000) Molecular portraits of human breast tumours. Nature 406: 747–752. - PubMed
-
- Andre F, Pusztai L (2006) Molecular classification of breast cancer: implications for selection of adjuvant chemotherapy. Nat Clin Pract Oncol 3: 621–632. - PubMed
-
- Lee H, Ormandy CJ (2012) Elf5, hormones and cell fate. Trends Endocrinol Metab in press. - PubMed
-
- Gutierrez-Hartmann A, Duval DL, Bradford AP (2007) ETS transcription factors in endocrine systems. Trends Endocrinol Metab 18: 150–158. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

