Chapter 3: Small molecules and disease
- PMID: 23300405
- PMCID: PMC3531289
- DOI: 10.1371/journal.pcbi.1002805
Chapter 3: Small molecules and disease
Abstract
"Big" molecules such as proteins and genes still continue to capture the imagination of most biologists, biochemists and bioinformaticians. "Small" molecules, on the other hand, are the molecules that most biologists, biochemists and bioinformaticians prefer to ignore. However, it is becoming increasingly apparent that small molecules such as amino acids, lipids and sugars play a far more important role in all aspects of disease etiology and disease treatment than we realized. This particular chapter focuses on an emerging field of bioinformatics called "chemical bioinformatics"--a discipline that has evolved to help address the blended chemical and molecular biological needs of toxicogenomics, pharmacogenomics, metabolomics and systems biology. In the following pages we will cover several topics related to chemical bioinformatics. First, a brief overview of some of the most important or useful chemical bioinformatic resources will be given. Second, a more detailed overview will be given on those particular resources that allow researchers to connect small molecules to diseases. This section will focus on describing a number of recently developed databases or knowledgebases that explicitly relate small molecules--either as the treatment, symptom or cause--to disease. Finally a short discussion will be provided on newly emerging software tools that exploit these databases as a means to discover new biomarkers or even new treatments for disease.
Conflict of interest statement
The author has declared that no competing interests exist.
Figures
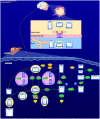

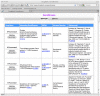
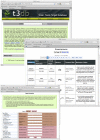
References
-
- Trujillo E, Davis C, Milner J (2006) Nutrigenomics, proteomics, metabolomics, and the practice of dietetics. J Am Diet Assoc 106: 403–413. - PubMed
-
- Feng X, Liu X, Luo Q, Liu BF (2008) Mass spectrometry in systems biology: an overview. Mass Spectrom Rev 27: 635–660. - PubMed
-
- Brown FK (1998) Chemoinformatics: what is it and how does it impact drug discovery. Annu Rep Med Chem 33: 375–384.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

