Chapter 5: Network biology approach to complex diseases
- PMID: 23300411
- PMCID: PMC3531284
- DOI: 10.1371/journal.pcbi.1002820
Chapter 5: Network biology approach to complex diseases
Abstract
Complex diseases are caused by a combination of genetic and environmental factors. Uncovering the molecular pathways through which genetic factors affect a phenotype is always difficult, but in the case of complex diseases this is further complicated since genetic factors in affected individuals might be different. In recent years, systems biology approaches and, more specifically, network based approaches emerged as powerful tools for studying complex diseases. These approaches are often built on the knowledge of physical or functional interactions between molecules which are usually represented as an interaction network. An interaction network not only reports the binary relationships between individual nodes but also encodes hidden higher level organization of cellular communication. Computational biologists were challenged with the task of uncovering this organization and utilizing it for the understanding of disease complexity, which prompted rich and diverse algorithmic approaches to be proposed. We start this chapter with a description of the general characteristics of complex diseases followed by a brief introduction to physical and functional networks. Next we will show how these networks are used to leverage genotype, gene expression, and other types of data to identify dysregulated pathways, infer the relationships between genotype and phenotype, and explain disease heterogeneity. We group the methods by common underlying principles and first provide a high level description of the principles followed by more specific examples. We hope that this chapter will give readers an appreciation for the wealth of algorithmic techniques that have been developed for the purpose of studying complex diseases as well as insight into their strengths and limitations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

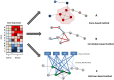
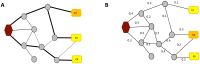
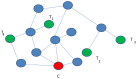
References
-
- Veenstra-Vanderweele J, Christian SL, Cook EH Jr (2004) Autism as a paradigmatic complex genetic disorder. Annu Rev Genomics Hum Genet 5: 379–405. - PubMed
-
- Schadt EE (2009) Molecular networks as sensors and drivers of common human diseases. Nature 461: 218–223. - PubMed
-
- Albert R (2005) Scale-free networks in cell biology. J Cell Sci 118: 4947–4957. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

