The underlying molecular and network level mechanisms in the evolution of robustness in gene regulatory networks
- PMID: 23300434
- PMCID: PMC3536627
- DOI: 10.1371/journal.pcbi.1002865
The underlying molecular and network level mechanisms in the evolution of robustness in gene regulatory networks
Abstract
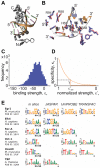
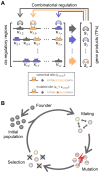
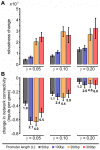
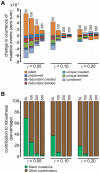
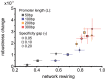
Gene regulatory networks show robustness to perturbations. Previous works identified robustness as an emergent property of gene network evolution but the underlying molecular mechanisms are poorly understood. We used a multi-tier modeling approach that integrates molecular sequence and structure information with network architecture and population dynamics. Structural models of transcription factor-DNA complexes are used to estimate relative binding specificities. In this model, mutations in the DNA cause changes on two levels: (a) at the sequence level in individual binding sites (modulating binding specificity), and (b) at the network level (creating and destroying binding sites). We used this model to dissect the underlying mechanisms responsible for the evolution of robustness in gene regulatory networks. Results suggest that in sparse architectures (represented by short promoters), a mixture of local-sequence and network-architecture level changes are exploited. At the local-sequence level, robustness evolves by decreasing the probabilities of both the destruction of existent and generation of new binding sites. Meanwhile, in highly interconnected architectures (represented by long promoters), robustness evolves almost entirely via network level changes, deleting and creating binding sites that modify the network architecture.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

