p12 tethers the murine leukemia virus pre-integration complex to mitotic chromosomes
- PMID: 23300449
- PMCID: PMC3531515
- DOI: 10.1371/journal.ppat.1003103
p12 tethers the murine leukemia virus pre-integration complex to mitotic chromosomes
Abstract
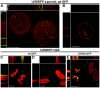
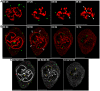
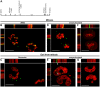
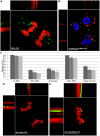
The p12 protein of the murine leukemia virus (MLV) is a constituent of the pre-integration complex (PIC) but its function in this complex remains unknown. We developed an imaging system to monitor MLV PIC trafficking in live cells. This allowed the visualization of PIC docking to mitotic chromosomes and its release upon exit from mitosis. Docking occurred concomitantly with nuclear envelope breakdown and was impaired for PICs of viruses with lethal p12 mutations. Insertion of a heterologous chromatin binding module into p12 of one of these mutants restored PICs attachment to the chromosomes and partially rescued virus replication. Capsid dissociated from wild type PICs in mitotic cells but remained associated with PICs harboring tethering-negative p12 mutants. Altogether, these results explain, in part, MLV restriction to dividing cells and reveal a role for p12 as a factor that tethers MLV PIC to mitotic chromosomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

