Genome-wide search for gene-gene interactions in colorectal cancer
- PMID: 23300701
- PMCID: PMC3530500
- DOI: 10.1371/journal.pone.0052535
Genome-wide search for gene-gene interactions in colorectal cancer
Abstract
Genome-wide association studies (GWAS) have successfully identified a number of single-nucleotide polymorphisms (SNPs) associated with colorectal cancer (CRC) risk. However, these susceptibility loci known today explain only a small fraction of the genetic risk. Gene-gene interaction (GxG) is considered to be one source of the missing heritability. To address this, we performed a genome-wide search for pair-wise GxG associated with CRC risk using 8,380 cases and 10,558 controls in the discovery phase and 2,527 cases and 2,658 controls in the replication phase. We developed a simple, but powerful method for testing interaction, which we term the Average Risk Due to Interaction (ARDI). With this method, we conducted a genome-wide search to identify SNPs showing evidence for GxG with previously identified CRC susceptibility loci from 14 independent regions. We also conducted a genome-wide search for GxG using the marginal association screening and examining interaction among SNPs that pass the screening threshold (p<10(-4)). For the known locus rs10795668 (10p14), we found an interacting SNP rs367615 (5q21) with replication p = 0.01 and combined p = 4.19×10(-8). Among the top marginal SNPs after LD pruning (n = 163), we identified an interaction between rs1571218 (20p12.3) and rs10879357 (12q21.1) (nominal combined p = 2.51×10(-6); Bonferroni adjusted p = 0.03). Our study represents the first comprehensive search for GxG in CRC, and our results may provide new insight into the genetic etiology of CRC.
Conflict of interest statement
Figures
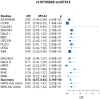

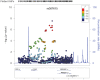
References
-
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, et al. (2007) A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nature genetics 39: 984–988 doi:10.1038/ng2085. - DOI - PubMed
-
- Zanke BW, Greenwood CMT, Rangrej J, Kustra R, Tenesa A, et al. (2007) Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nature genetics 39: 989–994 doi:10.1038/ng2089. - DOI - PubMed
-
- Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, et al. (2007) A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nature genetics 39: 1315–1317 doi:10.1038/ng.2007.18. - DOI - PubMed
-
- Tenesa A, Farrington SM, Prendergast JGD, Porteous ME, Walker M, et al. (2008) Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nature genetics 40: 631–637 doi:10.1038/ng.133. - DOI - PMC - PubMed
-
- Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, et al. (2008) Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nature genetics 40: 26–28 doi:10.1038/ng.2007.41. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
- U01 HG004438/HG/NHGRI NIH HHS/United States
- R01 AG014358/AG/NIA NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- K05 CA154337/CA/NCI NIH HHS/United States
- U01 CA074794/CA/NCI NIH HHS/United States
- R37 CA54281/CA/NCI NIH HHS/United States
- R01 CA48998/CA/NCI NIH HHS/United States
- R01 CA042182/CA/NCI NIH HHS/United States
- CA42182/CA/NCI NIH HHS/United States
- U01 CA122839/CA/NCI NIH HHS/United States
- CA-95-011/CA/NCI NIH HHS/United States
- R37 CA054281/CA/NCI NIH HHS/United States
- U19 CA148107/CA/NCI NIH HHS/United States
- R01 CA059045/CA/NCI NIH HHS/United States
- R01 CA60987/CA/NCI NIH HHS/United States
- R01 CA076366/CA/NCI NIH HHS/United States
- P01 CA 087969/CA/NCI NIH HHS/United States
- U01 HG004446/HG/NHGRI NIH HHS/United States
- P30 CA015704/CA/NCI NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- Z01 CP010200/ImNIH/Intramural NIH HHS/United States
- P01 CA055075/CA/NCI NIH HHS/United States
- P50 CA 127003/CA/NCI NIH HHS/United States
- N01 WH022110/WH/WHI NIH HHS/United States
- R01 137178/PHS HHS/United States
- CAPMC/ CIHR/Canada
- R01 CA048998/CA/NCI NIH HHS/United States
- U01 CA137088/CA/NCI NIH HHS/United States
- P01 CA 055075/CA/NCI NIH HHS/United States
- U01 HG 004438/HG/NHGRI NIH HHS/United States
- R01 CA063464/CA/NCI NIH HHS/United States
- P01 CA033619/CA/NCI NIH HHS/United States
- R01 CA060987/CA/NCI NIH HHS/United States
- U01 CA097735/CA/NCI NIH HHS/United States
- R01 CA63464/CA/NCI NIH HHS/United States
- U01 CA074783/CA/NCI NIH HHS/United States
- Z01 CP 010200/CP/NCI NIH HHS/United States
- P50 CA127003/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

