Acute hypersensitivity of pluripotent testicular cancer-derived embryonal carcinoma to low-dose 5-aza deoxycytidine is associated with global DNA Damage-associated p53 activation, anti-pluripotency and DNA demethylation
- PMID: 23300844
- PMCID: PMC3531428
- DOI: 10.1371/journal.pone.0053003
Acute hypersensitivity of pluripotent testicular cancer-derived embryonal carcinoma to low-dose 5-aza deoxycytidine is associated with global DNA Damage-associated p53 activation, anti-pluripotency and DNA demethylation
Abstract
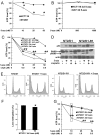
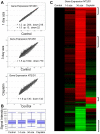
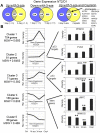
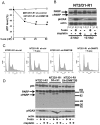
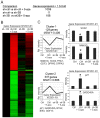
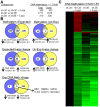
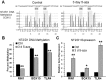
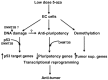
Human embryonal carcinoma (EC) cells are the stem cells of nonseminoma testicular germ cells tumors (TGCTs) and share remarkable similarities to human embryonic stem (ES) cells. In prior work we found that EC cells are hypersensitive to low nanomolar doses of 5-aza deoxycytidine (5-aza) and that this hypersensitivity partially depended on unusually high levels of the DNA methyltransferase, DNMT3B. We show here that low-dose 5-aza treatment results in DNA damage and induction of p53 in NT2/D1 cells. In addition, low-dose 5-aza results in global and gene specific promoter DNA hypomethylation. Low-dose 5-aza induces a p53 transcriptional signature distinct from that induced with cisplatin in NT2/D1 cells and also uniquely downregulates genes associated with pluripotency including NANOG, SOX2, GDF3 and Myc target genes. Changes in the p53 and pluripotency signatures with 5-aza were to a large extent dependent on high levels of DNMT3B. In contrast to the majority of p53 target genes upregulated by 5-aza that did not show DNA hypomethylation, several other genes induced with 5-aza had corresponding decreases in promoter methylation. These genes include RIN1, SOX15, GPER, and TLR4 and are novel candidate tumors suppressors in TGCTs. Our studies suggest that the hypersensitivity of NT2/D1 cells to low-dose 5-aza is multifactorial and involves the combined activation of p53 targets, repression of pluripotency genes, and activation of genes repressed by DNA methylation. Low-dose 5-aza therapy may be a general strategy to treat those tumors that are sustained by cells with embryonic stem-like properties.GEO NUMBER FOR THE MICROARRAY DATA: GSE42647.
Conflict of interest statement
Figures
References
-
- Koychev D, Oechsle K, Bokemeyer C, Honecker F (2011) Treatment of patients with relapsed and/or cisplatin-refractory metastatic germ cell tumours: an update. Int J Androl 34: e266–273. - PubMed
-
- Efstathiou E, Logothetis CJ (2006) Review of late complications of treatment and late relapse in testicular cancer. J Natl Compr Canc Netw 4: 1059–1070. - PubMed
-
- Kristensen DM, Sonne SB, Ottesen AM, Perrett RM, Nielsen JE, et al. (2008) Origin of pluripotent germ cell tumours: the role of microenvironment during embryonic development. Mol Cell Endocrinol 288: 111–118. - PubMed
-
- Clark AT (2007) The stem cell identity of testicular cancer. Stem Cell Rev 3: 49–59. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

