The P body protein Dcp1a is hyper-phosphorylated during mitosis
- PMID: 23300942
- PMCID: PMC3534667
- DOI: 10.1371/journal.pone.0049783
The P body protein Dcp1a is hyper-phosphorylated during mitosis
Abstract
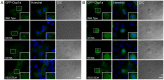
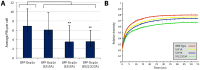
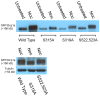
Processing bodies (PBs) are non-membranous cytoplasmic structures found in all eukaryotes. Many of their components such as the Dcp1 and Dcp2 proteins are highly conserved. Using live-cell imaging we found that PB structures disassembled as cells prepared for cell division, and then began to reassemble during the late stages of cytokinesis. During the cell cycle and as cells passed through S phase, PB numbers increased. However, there was no memory of PB numbers between mother and daughter cells. Examination of hDcp1a and hDcp1b proteins by electrophoresis in mitotic cell extracts showed a pronounced slower migrating band, which was caused by hyper-phosphorylation of the protein. We found that hDcp1a is a phospho-protein during interphase that becomes hyper-phosphorylated in mitotic cells. Using truncations of hDcp1a we localized the region important for hyper-phosphorylation to the center of the protein. Mutational analysis demonstrated the importance of serine 315 in the hyper-phosphorylation process, while other serine residues tested had a minor affect. Live-cell imaging demonstrated that serine mutations in other regions of the protein affected the dynamics of hDcp1a association with the PB structure. Our work demonstrates the control of PB dynamics during the cell cycle via phosphorylation.
Conflict of interest statement
Figures
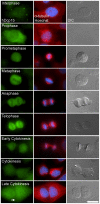
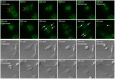

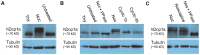
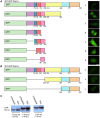
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

