Functional analysis of the ComK protein of Bacillus coagulans
- PMID: 23301076
- PMCID: PMC3536758
- DOI: 10.1371/journal.pone.0053471
Functional analysis of the ComK protein of Bacillus coagulans
Erratum in
- PLoS One. 2013;8(2). doi:10.1371/annotation/16c64f49-b803-458d-8c8c-fa22a9c649b2. van Hartskamp, Mariska [added]
Abstract
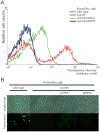
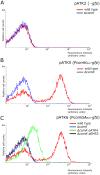
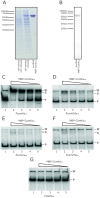
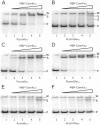
The genes for DNA uptake and recombination in Bacilli are commonly regulated by the transcriptional factor ComK. We have identified a ComK homologue in Bacillus coagulans, an industrial relevant organism that is recalcitrant for transformation. Introduction of B. coagulans comK gene under its own promoter region into Bacillus subtilis comK strain results in low transcriptional induction of the late competence gene comGA, but lacking bistable expression. The promoter regions of B. coagulans comK and the comGA genes are recognized in B. subtilis and expression from these promoters is activated by B. subtilis ComK. Purified ComK protein of B. coagulans showed DNA-binding ability in gel retardation assays with B. subtilis- and B. coagulans-derived probes. These experiments suggest that the function of B. coagulans ComK is similar to that of ComK of B. subtilis. When its own comK is overexpressed in B. coagulans the comGA gene expression increases 40-fold, while the expression of another late competence gene, comC is not elevated and no reproducible DNA-uptake could be observed under these conditions. Our results demonstrate that B. coagulans ComK can recognize several B.subtilis comK-responsive elements, and vice versa, but indicate that the activation of the transcription of complete sets of genes coding for a putative DNA uptake apparatus in B. coagulans might differ from that of B. subtilis.
Conflict of interest statement
Figures

Similar articles
-
The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix.Genes Dev. 1998 May 15;12(10):1539-50. doi: 10.1101/gad.12.10.1539. Genes Dev. 1998. PMID: 9585513 Free PMC article.
-
comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis.Mol Microbiol. 1995 Feb;15(3):455-62. doi: 10.1111/j.1365-2958.1995.tb02259.x. Mol Microbiol. 1995. PMID: 7783616
-
Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach.Nucleic Acids Res. 2002 Dec 15;30(24):5517-28. doi: 10.1093/nar/gkf698. Nucleic Acids Res. 2002. PMID: 12490720 Free PMC article.
-
Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond.World J Microbiol Biotechnol. 2018 Sep 10;34(10):145. doi: 10.1007/s11274-018-2531-7. World J Microbiol Biotechnol. 2018. PMID: 30203131 Review.
-
Ubiquitous late competence genes in Bacillus species indicate the presence of functional DNA uptake machineries.Environ Microbiol. 2009 Aug;11(8):1911-22. doi: 10.1111/j.1462-2920.2009.01937.x. Epub 2009 May 14. Environ Microbiol. 2009. PMID: 19453701 Review.
Cited by
-
In vivo selection of sfGFP variants with improved and reliable functionality in industrially important thermophilic bacteria.Biotechnol Biofuels. 2018 Jan 17;11:8. doi: 10.1186/s13068-017-1008-5. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29371884 Free PMC article.
-
Genomic analysis of thermophilic Bacillus coagulans strains: efficient producers for platform bio-chemicals.Sci Rep. 2014 Jan 29;4:3926. doi: 10.1038/srep03926. Sci Rep. 2014. PMID: 24473268 Free PMC article.
-
Multitasking functions of bacterial extracellular DNA in biofilms.J Bacteriol. 2024 Apr 18;206(4):e0000624. doi: 10.1128/jb.00006-24. Epub 2024 Mar 6. J Bacteriol. 2024. PMID: 38445859 Free PMC article. Review.
-
ComFB, a new widespread family of c-di-NMP receptor proteins.bioRxiv [Preprint]. 2024 Nov 10:2024.11.10.622515. doi: 10.1101/2024.11.10.622515. bioRxiv. 2024. PMID: 39574629 Free PMC article. Preprint.
-
Natural DNA Transformation Is Functional in Lactococcus lactis subsp. cremoris KW2.Appl Environ Microbiol. 2017 Aug 1;83(16):e01074-17. doi: 10.1128/AEM.01074-17. Print 2017 Aug 15. Appl Environ Microbiol. 2017. PMID: 28625996 Free PMC article.
References
-
- Claverys JP, Prudhomme M, Martin B (2006) Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu Rev Microbiol 60: 451–475. - PubMed
-
- Martin B, Quentin Y, Fichant G, Claverys JP (2006) Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol 14(8): 339–345. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

