Functional genomics of a generalist parasitic plant: laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression
- PMID: 23302495
- PMCID: PMC3636017
- DOI: 10.1186/1471-2229-13-9
Functional genomics of a generalist parasitic plant: laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression
Abstract
Background: Orobanchaceae is the only plant family with members representing the full range of parasitic lifestyles plus a free-living lineage sister to all parasitic lineages, Lindenbergia. A generalist member of this family, and an important parasitic plant model, Triphysaria versicolor regularly feeds upon a wide range of host plants. Here, we compare de novo assembled transcriptomes generated from laser micro-dissected tissues at the host-parasite interface to uncover details of the largely uncharacterized interaction between parasitic plants and their hosts.
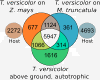
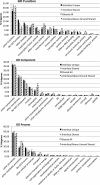
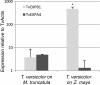
Results: The interaction of Triphysaria with the distantly related hosts Zea mays and Medicago truncatula reveals dramatic host-specific gene expression patterns. Relative to above ground tissues, gene families are disproportionally represented at the interface including enrichment for transcription factors and genes of unknown function. Quantitative Real-Time PCR of a T. versicolor β-expansin shows strong differential (120x) upregulation in response to the monocot host Z. mays; a result that is concordant with our read count estimates. Pathogenesis-related proteins, other cell wall modifying enzymes, and orthologs of genes with unknown function (annotated as such in sequenced plant genomes) are among the parasite genes highly expressed by T. versicolor at the parasite-host interface.
Conclusions: Laser capture microdissection makes it possible to sample the small region of cells at the epicenter of parasite host interactions. The results of our analysis suggest that T. versicolor's generalist strategy involves a reliance on overlapping but distinct gene sets, depending upon the host plant it is parasitizing. The massive upregulation of a T. versicolor β-expansin is suggestive of a mechanism for parasite success on grass hosts. In this preliminary study of the interface transcriptomes, we have shown that T. versicolor, and the Orobanchaceae in general, provide excellent opportunities for the characterization of plant genes with unknown functions.
Figures

References
-
- Ejeta G. In: Integrating New Technologies for Striga Control: Towards Ending the Witch-hunt. Gressel J, editor. Singapore: World Scientific Publishing Co; 2007. The Striga scourge in Africa: a growing pandemic; pp. 3–16.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

