How insulin engages its primary binding site on the insulin receptor
- PMID: 23302862
- PMCID: PMC3793637
- DOI: 10.1038/nature11781
How insulin engages its primary binding site on the insulin receptor
Abstract
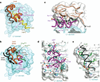
Insulin receptor signalling has a central role in mammalian biology, regulating cellular metabolism, growth, division, differentiation and survival. Insulin resistance contributes to the pathogenesis of type 2 diabetes mellitus and the onset of Alzheimer's disease; aberrant signalling occurs in diverse cancers, exacerbated by cross-talk with the homologous type 1 insulin-like growth factor receptor (IGF1R). Despite more than three decades of investigation, the three-dimensional structure of the insulin-insulin receptor complex has proved elusive, confounded by the complexity of producing the receptor protein. Here we present the first view, to our knowledge, of the interaction of insulin with its primary binding site on the insulin receptor, on the basis of four crystal structures of insulin bound to truncated insulin receptor constructs. The direct interaction of insulin with the first leucine-rich-repeat domain (L1) of insulin receptor is seen to be sparse, the hormone instead engaging the insulin receptor carboxy-terminal α-chain (αCT) segment, which is itself remodelled on the face of L1 upon insulin binding. Contact between insulin and L1 is restricted to insulin B-chain residues. The αCT segment displaces the B-chain C-terminal β-strand away from the hormone core, revealing the mechanism of a long-proposed conformational switch in insulin upon receptor engagement. This mode of hormone-receptor recognition is novel within the broader family of receptor tyrosine kinases. We support these findings by photo-crosslinking data that place the suggested interactions into the context of the holoreceptor and by isothermal titration calorimetry data that dissect the hormone-insulin receptor interface. Together, our findings provide an explanation for a wealth of biochemical data from the insulin receptor and IGF1R systems relevant to the design of therapeutic insulin analogues.
Figures
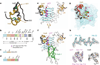

Comment in
-
Structural biology: Insulin meets its receptor.Nature. 2013 Jan 10;493(7431):171-2. doi: 10.1038/493171a. Nature. 2013. PMID: 23302856 No abstract available.
-
Milestone: meet and greet-insulin and receptor interaction unraveled.Nat Rev Endocrinol. 2013 Mar;9(3):131. doi: 10.1038/nrendo.2013.23. Epub 2013 Feb 5. Nat Rev Endocrinol. 2013. PMID: 23381032 No abstract available.
References
-
- Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nature Rev. Mol. Cell Biol. 2006;7:85–96. - PubMed
-
- Cohen P. The twentieth century struggle to decipher insulin signalling. Nature Rev. Mol. Cell Biol. 2006;7:867–873. - PubMed
-
- Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nature Rev. Cancer. 2012;12:159–169. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

