Clostridium difficile TcdC protein binds four-stranded G-quadruplex structures
- PMID: 23303781
- PMCID: PMC3575817
- DOI: 10.1093/nar/gks1448
Clostridium difficile TcdC protein binds four-stranded G-quadruplex structures
Abstract
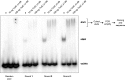
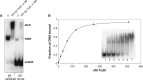
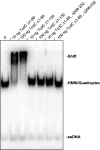
Clostridium difficile infections are increasing worldwide due to emergence of virulent strains. Infections can result in diarrhea and potentially fatal pseudomembranous colitis. The main virulence factors of C. difficile are clostridial toxins TcdA and TcdB. Transcription of the toxins is positively regulated by the sigma factor TcdR. Negative regulation is believed to occur through TcdC, a proposed anti-sigma factor. Here, we describe the biochemical properties of TcdC to understand the mechanism of TcdC action. Bioinformatic analysis of the TcdC protein sequence predicted the presence of a hydrophobic stretch [amino acids (aa) 30-50], a potential dimerization domain (aa 90-130) and a C-terminal oligonucleotide-binding fold. Gel filtration chromatography of two truncated recombinant TcdC proteins (TcdCΔ1-89 and TcdCΔ1-130) showed that the domain between aa 90 and 130 is involved in dimerization. Binding of recombinant TcdC to single-stranded DNA was studied using a single-stranded Systematic Evolution of Ligands by Exponential enrichment approach. This involved specific binding of single-stranded DNA sequences from a pool of random oligonucleotides, as monitored by electrophoretic-mobility shift assay. Analysis of the oligonucleotides bound showed that the oligonucleotide-binding fold domain of TcdC can bind specifically to DNA folded into G-quadruplex structures containing repetitive guanine nucleotides forming a four-stranded structure. In summary, we provide evidence for DNA binding of TcdC, which suggests an alternative function for this proposed anti-sigma factor.
Figures


References
-
- Goorhuis A., Bakker D., Corver J., Debast S.B., Harmanus C., Notermans D.W., Bergwerff A.A., Dekker F.W., Kuijper E.J. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 2008;47:1162–1170. - PubMed
-
- Kuijper E.J., Barbut F., Brazier J.S., Kleinkauf N., Eckmanns T., Lambert M.L., Drudy D., Fitzpatrick F., Wiuff C., Brown D.J., et al. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro. Surveill. 2008;13:pii: 18942. - PubMed
-
- McDonald L.C., Killgore G.E., Thompson A., Owens R.C., Jr, Kazakova S.V., Sambol S.P., Johnson S., Gerding D.N. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 2005;353:2433–2441. - PubMed
-
- Rupnik M., Wilcox M.H., Gerding D.N. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 2009;7:526–536. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

