Functional characterization of motif sequences under purifying selection
- PMID: 23303791
- PMCID: PMC3575792
- DOI: 10.1093/nar/gks1456
Functional characterization of motif sequences under purifying selection
Abstract
Diverse life forms are driven by the evolution of gene regulatory programs including changes in regulator proteins and cis-regulatory elements. Alterations of cis-regulatory elements are likely to dominate the evolution of the gene regulatory networks, as they are subjected to smaller selective constraints compared with proteins and hence may evolve quickly to adapt the environment. Prior studies on cis-regulatory element evolution focus primarily on sequence substitutions of known transcription factor-binding motifs. However, evolutionary models for the dynamics of motif occurrence are relatively rare, and comprehensive characterization of the evolution of all possible motif sequences has not been pursued. In the present study, we propose an algorithm to estimate the strength of purifying selection of a motif sequence based on an evolutionary model capturing the birth and death of motif occurrences on promoters. We term this measure as the 'evolutionary retention coefficient', as it is related yet distinct from the canonical definition of selection coefficient in population genetics. Using this algorithm, we estimate and report the evolutionary retention coefficients of all possible 10-nucleotide sequences from the aligned promoter sequences of 27 748. orthologous gene families in 34 mammalian species. Intriguingly, the evolutionary retention coefficients of motifs are intimately associated with their functional relevance. Top-ranking motifs (sorted by evolutionary retention coefficients) are significantly enriched with transcription factor-binding sequences according to the curated knowledge from the TRANSFAC database and the ChIP-seq data generated from the ENCODE Consortium. Moreover, genes harbouring high-scoring motifs on their promoters retain significantly coherent expression profiles, and those genes are over-represented in the functional classes involved in gene regulation. The validation results reveal the dependencies between natural selection and functions of cis-regulatory elements and shed light on the evolution of gene regulatory networks.
Figures
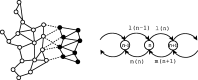
 denotes the count of motif occurrence on a promoter.
denotes the count of motif occurrence on a promoter.  and
and  denote the birth and death rates emanating from state
denote the birth and death rates emanating from state  .
.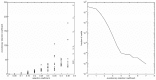
 10-mer sequences. The probabilities are displayed in a log scale.
10-mer sequences. The probabilities are displayed in a log scale.
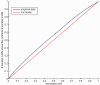
 in equation 10]. The red curve shows the CDF of a uniform distribution [
in equation 10]. The red curve shows the CDF of a uniform distribution [ ].
].
Similar articles
-
A fast weak motif-finding algorithm based on community detection in graphs.BMC Bioinformatics. 2013 Jul 17;14:227. doi: 10.1186/1471-2105-14-227. BMC Bioinformatics. 2013. PMID: 23865838 Free PMC article.
-
An integrative and applicable phylogenetic footprinting framework for cis-regulatory motifs identification in prokaryotic genomes.BMC Genomics. 2016 Aug 9;17:578. doi: 10.1186/s12864-016-2982-x. BMC Genomics. 2016. PMID: 27507169 Free PMC article.
-
Differential motif enrichment analysis of paired ChIP-seq experiments.BMC Genomics. 2014 Sep 2;15(1):752. doi: 10.1186/1471-2164-15-752. BMC Genomics. 2014. PMID: 25179504 Free PMC article.
-
The Identification of Cis-Regulatory Sequence Motifs in Gene Promoters Based on SNP Information.Methods Mol Biol. 2016;1482:31-47. doi: 10.1007/978-1-4939-6396-6_3. Methods Mol Biol. 2016. PMID: 27557759
-
An algorithmic perspective of de novo cis-regulatory motif finding based on ChIP-seq data.Brief Bioinform. 2018 Sep 28;19(5):1069-1081. doi: 10.1093/bib/bbx026. Brief Bioinform. 2018. PMID: 28334268 Review.
Cited by
-
Prediction and Validation of Transcription Factors Modulating the Expression of Sestrin3 Gene Using an Integrated Computational and Experimental Approach.PLoS One. 2016 Jul 28;11(7):e0160228. doi: 10.1371/journal.pone.0160228. eCollection 2016. PLoS One. 2016. PMID: 27466818 Free PMC article.
References
-
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. - PubMed
-
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. - PubMed
-
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory motifs. Nature. 2003;423:241–254. - PubMed
-
- Siepel A, Haussler D. Combining phylogenetic and hidden Markov models in biosequence analysis. J. Comput. Biol. 2004;11:413–428. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

