Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures
- PMID: 23303917
- PMCID: PMC3584537
- DOI: 10.1105/tpc.112.105643
Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures
Abstract
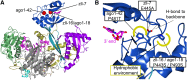
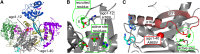
RNA silencing refers to a collection of gene regulatory mechanisms that use small RNAs for sequence specific repression. These mechanisms rely on ARGONAUTE (AGO) proteins that directly bind small RNAs and thereby constitute the central component of the RNA-induced silencing complex (RISC). AGO protein function has been probed extensively by mutational analyses, particularly in plants where large allelic series of several AGO proteins have been isolated. Structures of entire human and yeast AGO proteins have only very recently been obtained, and they allow more precise analyses of functional consequences of mutations obtained by forward genetics. To a large extent, these analyses support current models of regions of particular functional importance of AGO proteins. Interestingly, they also identify previously unrecognized parts of AGO proteins with profound structural and functional importance and provide the first hints at structural elements that have important functions specific to individual AGO family members. A particularly important outcome of the analysis concerns the evidence for existence of Gly-Trp (GW) repeat interactors of AGO proteins acting in the plant microRNA pathway. The parallel analysis of AGO structures and plant AGO mutations also suggests that such interactions with GW proteins may be a determinant of whether an endonucleolytically competent RISC is formed.
Figures





References
-
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

