Accelerated high-yield generation of limb-innervating motor neurons from human stem cells
- PMID: 23303937
- PMCID: PMC3711539
- DOI: 10.1523/JNEUROSCI.0906-12.2013
Accelerated high-yield generation of limb-innervating motor neurons from human stem cells
Abstract
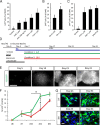
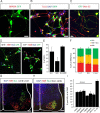
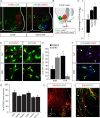
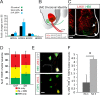
Human pluripotent stem cells are a promising source of differentiated cells for developmental studies, cell transplantation, disease modeling, and drug testing. However, their widespread use even for intensely studied cell types like spinal motor neurons is hindered by the long duration and low yields of existing protocols for in vitro differentiation and by the molecular heterogeneity of the populations generated. We report a combination of small molecules that within 3 weeks induce motor neurons at up to 50% abundance and with defined subtype identities of relevance to neurodegenerative disease. Despite their accelerated differentiation, motor neurons expressed combinations of HB9, ISL1, and column-specific markers that mirror those observed in vivo in human embryonic spinal cord. They also exhibited spontaneous and induced activity, and projected axons toward muscles when grafted into developing chick spinal cord. Strikingly, this novel protocol preferentially generates motor neurons expressing markers of limb-innervating lateral motor column motor neurons (FOXP1(+)/LHX3(-)). Access to high-yield cultures of human limb-innervating motor neuron subtypes will facilitate in-depth study of motor neuron subtype-specific properties, disease modeling, and development of large-scale cell-based screening assays.
Figures

Comment in
-
Induced pluripotent stem cells and motor neuron disease: toward an era of individualized medicine.J Neurosci. 2013 May 15;33(20):8587-9. doi: 10.1523/JNEUROSCI.1062-13.2013. J Neurosci. 2013. PMID: 23678103 Free PMC article. No abstract available.
References
-
- Arber S, Han B, Mendelsohn M, Smith M, Jessell TM, Sockanathan S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
