Top down proteomics of human membrane proteins from enriched mitochondrial fractions
- PMID: 23305238
- PMCID: PMC3565750
- DOI: 10.1021/ac3031527
Top down proteomics of human membrane proteins from enriched mitochondrial fractions
Abstract
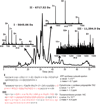
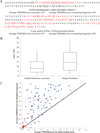
The interrogation of intact integral membrane proteins has long been a challenge for biological mass spectrometry. Here, we demonstrate the application of top down mass spectrometry to whole membrane proteins below 60 kDa with up to 8 transmembrane helices. Analysis of enriched mitochondrial membrane preparations from human cells yielded identification of 83 integral membrane proteins, along with 163 membrane-associated or soluble proteins, with a median q value of 3 × 10(-10). An analysis of matching fragment ions demonstrated that significantly more fragment ions were found within transmembrane domains than would be expected based upon the observed protein sequence. In total, 46 proteins from the complexes of oxidative phosphorylation were identified which exemplifies the increasing ability of top down proteomics to provide extensive coverage in a biological network.
Figures



References
-
- Siuti N, Roth MJ, Mizzen CA, Kelleher NL, Pesavento JJ. J. Proteome Res. 2006;5:233–239. - PubMed
-
- Resemann A, Wunderlich D, Rothbauer U, Warscheid B, Leonhardt H, Fuchser J, Kuhlmann K, Suckau D. Anal. Chem. 2010;82:3283–3292. - PubMed
-
- Meng FY, Cargile BJ, Patrie SM, Johnson JR, McLoughlin SM, Kelleher NL. Anal. Chem. 2002;74:2923–2929. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

