Polyproline and triple helix motifs in host-pathogen recognition
- PMID: 23305370
- PMCID: PMC3707005
- DOI: 10.2174/138920312804871157
Polyproline and triple helix motifs in host-pathogen recognition
Abstract
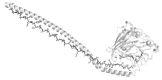
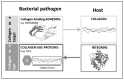
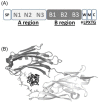
Secondary structure elements often mediate protein-protein interactions. Despite their low abundance in folded proteins, polyproline II (PPII) and its variant, the triple helix, are frequently involved in protein-protein interactions, likely due to their peculiar propensity to be solvent-exposed. We here review the role of PPII and triple helix in mediating hostpathogen interactions, with a particular emphasis to the structural aspects of these processes. After a brief description of the basic structural features of these elements, examples of host-pathogen interactions involving these motifs are illustrated. Literature data suggest that the role played by PPII motif in these processes is twofold. Indeed, PPII regions may directly mediate interactions between proteins of the host and the pathogen. Alternatively, PPII may act as structural spacers needed for the correct positioning of the elements needed for adhesion and infectivity. Recent investigations have highlighted that collagen triple helix is also a common target for bacterial adhesins. Although structural data on complexes between adhesins and collagen models are rather limited, experimental and theoretical studies have unveiled some interesting clues of the recognition process. Interestingly, very recent data show that not only is the triple helix used by pathogens as a target in the host-pathogen interaction but it may also act as a bait in these processes since bacterial proteins containing triple helix regions have been shown to interact with host proteins. As both PPII and triple helix expose several main chain non-satisfied hydrogen bond acceptors and donors, both elements are highly solvated. The preservation of the solvation state of both PPII and triple helix upon protein-protein interaction is an emerging aspect that will be here thoroughly discussed.
Figures



Similar articles
-
Polyproline-II helix in proteins: structure and function.J Mol Biol. 2013 Jun 26;425(12):2100-32. doi: 10.1016/j.jmb.2013.03.018. Epub 2013 Mar 16. J Mol Biol. 2013. PMID: 23507311 Review.
-
Intramolecular binding of a proximal PPII helix to an SH3 domain in the fusion protein SH3Hck : PPIIhGAP.Cell Biochem Biophys. 2001;35(2):115-26. doi: 10.1385/cbb:35:2:115. Cell Biochem Biophys. 2001. PMID: 11892787
-
Properties of polyproline II, a secondary structure element implicated in protein-protein interactions.Proteins. 2005 Mar 1;58(4):880-92. doi: 10.1002/prot.20327. Proteins. 2005. PMID: 15657931
-
Host-guest scale of left-handed polyproline II helix formation.Proteins. 2003 Oct 1;53(1):68-75. doi: 10.1002/prot.10477. Proteins. 2003. PMID: 12945050
-
The structure of "unstructured" regions in peptides and proteins: role of the polyproline II helix in protein folding and recognition.Biopolymers. 2005;80(2-3):179-85. doi: 10.1002/bip.20227. Biopolymers. 2005. PMID: 15700296 Review.
Cited by
-
The crystal structure of the streptococcal collagen-like protein 2 globular domain from invasive M3-type group A Streptococcus shows significant similarity to immunomodulatory HIV protein gp41.J Biol Chem. 2014 Feb 21;289(8):5122-33. doi: 10.1074/jbc.M113.523597. Epub 2013 Dec 19. J Biol Chem. 2014. PMID: 24356966 Free PMC article.
-
The Proline/Glycine-Rich Region of the Biofilm Adhesion Protein Aap Forms an Extended Stalk that Resists Compaction.J Mol Biol. 2017 Jan 20;429(2):261-279. doi: 10.1016/j.jmb.2016.11.017. Epub 2016 Nov 25. J Mol Biol. 2017. PMID: 27890783 Free PMC article.
-
Structure of human Aichi virus and implications for receptor binding.Nat Microbiol. 2016 Sep 5;1(11):16150. doi: 10.1038/nmicrobiol.2016.150. Nat Microbiol. 2016. PMID: 27595320
-
Staphylococcus aureus SspA (V8 protease): New skin pathogenesis insights into an old enzyme.PLoS Pathog. 2025 Apr 24;21(4):e1013048. doi: 10.1371/journal.ppat.1013048. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40273042 Free PMC article. No abstract available.
-
Transcription profile of Trichophyton rubrum conidia grown on keratin reveals the induction of an adhesin-like protein gene with a tandem repeat pattern.BMC Genomics. 2016 Mar 18;17:249. doi: 10.1186/s12864-016-2567-8. BMC Genomics. 2016. PMID: 26993619 Free PMC article.
References
-
- Richardson JS. The anatomy and taxonomy of protein structure. Adv. Prot. Chem. 1981;34:167–339. - PubMed
-
- Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. - PubMed
-
- Unger R, Sussman JL. The importance of short structural motifs in protein structure analysis. J. Comp.-Aid. Mol. Des. 1993;7:457–472. - PubMed
-
- Brenner SE, Chothia C, Hubbard TJ. Population statistics of protein structures: Lessons from structural classifications. Curr. Opin. Struct. Biol. 1997;7:369–376. - PubMed
-
- De Simone A, Berisio R, Zagari A, Vitagliano L. Limited tendency of alpha-helical residues to form disulfide bridges: A structural explanation. J. Pept. Sci. 2006;12:740–747. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
