Streamlined protocol for mRNA display
- PMID: 23305392
- PMCID: PMC3666848
- DOI: 10.1021/co300135r
Streamlined protocol for mRNA display
Abstract
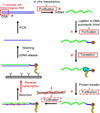
mRNA display is a powerful method for in vitro directed evolution of polypeptides, but its time-consuming, technically demanding nature has hindered its widespread use. We present a streamlined protocol in which lengthy mRNA purification steps are replaced with faster precipitation and ultrafiltration alternatives; additionally, other purification steps are entirely eliminated by using a reconstituted translation system and by performing reverse transcription after selection, which also protects input polypeptides from thermal denaturation. We tested this procedure by performing affinity selection against Her2 using binary libraries containing a nonspecific designed ankyrin repeat protein (DARPin) doped with a Her2-binding DARPin (dopant fraction ranging from 1:10 to 1:10 000). The Her2-binding DARPin was recovered in all cases, with an enrichment factor of up to 2 orders of magnitude per selection round. The time required for 1 round is reduced from ∼4-7 days to 2 days with our protocol, thus simplifying and accelerating mRNA display experiments.
Figures

References
-
- Dreier B, Plückthun A. Ribosome display: a technology for selecting and evolving proteins from large libraries. Methods Mol. Biol. 2011;687:283–306. - PubMed
-
- Keefe AD. Protein selection using mRNA display. Curr. Protoc. Mol. Biol. :24.5.1–24.5.34. - PubMed
-
- Miyamoto-Sato E, Fujimori S, Ishizaka M, Hirai N, Masuoka K, Saito R, Ozawa Y, Hino K, Washio T, Tomita M, Yamashita T, Oshikubo T, Akasaka H, Sugiyama J, Matsumoto Y, Yanagawa H. A comprehensive resource of interacting protein regions for refining human transcription factor networks. PLoS One. 2010;5:e9289. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

