Ensemble analysis of primary microRNA structure reveals an extensive capacity to deform near the Drosha cleavage site
- PMID: 23305493
- PMCID: PMC3565094
- DOI: 10.1021/bi301452a
Ensemble analysis of primary microRNA structure reveals an extensive capacity to deform near the Drosha cleavage site
Abstract
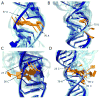
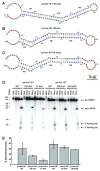
Most noncoding RNAs function properly only when folded into complex three-dimensional (3D) structures, but the experimental determination of these structures remains challenging. Understanding of primary microRNA (miRNA) maturation is currently limited by a lack of determined structures for nonprocessed forms of the RNA. SHAPE chemistry efficiently determines RNA secondary structural information with single-nucleotide resolution, providing constraints suitable for input into MC-Pipeline for refinement of 3D structure models. Here we combine these approaches to analyze three structurally diverse primary microRNAs, revealing deviations from canonical double-stranded RNA structure in the stem adjacent to the Drosha cut site for all three. The necessity of these deformable sites for efficient processing is demonstrated through Drosha processing assays. The structure models generated herein support the hypothesis that deformable sequences spaced roughly once per turn of A-form helix, created by noncanonical structure elements, combine with the necessary single-stranded RNA-double-stranded RNA junction to define the correct Drosha cleavage site.
Figures




Similar articles
-
A central role for the primary microRNA stem in guiding the position and efficiency of Drosha processing of a viral pri-miRNA.RNA. 2014 Jul;20(7):1068-77. doi: 10.1261/rna.044537.114. Epub 2014 May 22. RNA. 2014. PMID: 24854622 Free PMC article.
-
Genome-wide Mapping of DROSHA Cleavage Sites on Primary MicroRNAs and Noncanonical Substrates.Mol Cell. 2017 Apr 20;66(2):258-269.e5. doi: 10.1016/j.molcel.2017.03.013. Mol Cell. 2017. PMID: 28431232
-
Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha.EMBO J. 2005 Jan 12;24(1):138-48. doi: 10.1038/sj.emboj.7600491. Epub 2004 Nov 25. EMBO J. 2005. PMID: 15565168 Free PMC article.
-
The role of the precursor structure in the biogenesis of microRNA.Cell Mol Life Sci. 2011 Sep;68(17):2859-71. doi: 10.1007/s00018-011-0726-2. Epub 2011 May 24. Cell Mol Life Sci. 2011. PMID: 21607569 Free PMC article. Review.
-
Regulation of primary microRNA processing.FEBS Lett. 2018 Jun;592(12):1980-1996. doi: 10.1002/1873-3468.13067. Epub 2018 May 8. FEBS Lett. 2018. PMID: 29683487 Review.
Cited by
-
Inhibition of MiR-92a May Protect Endothelial Cells After Acute Myocardial Infarction in Rats: Role of KLF2/4.Med Sci Monit. 2016 Jul 14;22:2451-62. doi: 10.12659/msm.897266. Med Sci Monit. 2016. PMID: 27411964 Free PMC article.
-
Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.Mol Cell. 2020 May 7;78(3):411-422.e4. doi: 10.1016/j.molcel.2020.02.016. Epub 2020 Mar 27. Mol Cell. 2020. PMID: 32220646 Free PMC article.
-
Engineering double-stranded RNA binding activity into the Drosha double-stranded RNA binding domain results in a loss of microRNA processing function.PLoS One. 2017 Aug 8;12(8):e0182445. doi: 10.1371/journal.pone.0182445. eCollection 2017. PLoS One. 2017. PMID: 28792523 Free PMC article.
-
Deformability in the cleavage site of primary microRNA is not sensed by the double-stranded RNA binding domains in the microprocessor component DGCR8.Proteins. 2015 Jun;83(6):1165-79. doi: 10.1002/prot.24810. Epub 2015 Apr 28. Proteins. 2015. PMID: 25851436 Free PMC article.
-
Elucidating the Role of Microprocessor Protein DGCR8 in Bending RNA Structures.Biophys J. 2020 Dec 15;119(12):2524-2536. doi: 10.1016/j.bpj.2020.10.038. Epub 2020 Nov 13. Biophys J. 2020. PMID: 33189689 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

