Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast
- PMID: 23305949
- PMCID: PMC3594648
- DOI: 10.1016/j.dnarep.2012.12.004
Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast
Abstract
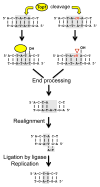
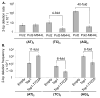
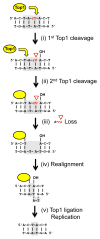
Topoisomerase 1 (Top1) resolves transcription-associated supercoils by generating transient single-strand breaks in DNA. Top1 activity in yeast is a major source of transcription-associated mutagenesis, generating a distinctive mutation signature characterized by deletions in short, tandem repeats. A similar signature is associated with the persistence of ribonucleoside monophosphates (rNMPs) in DNA, and it also depends on Top1 activity. There is only partial overlap, however, between Top1-dependent deletion hotspots identified in highly transcribed DNA and those associated with rNMPs, suggesting the existence of both rNMP-dependent and rNMP-independent events. Here, we present genetic studies confirming that there are two distinct types of hotspots. Data suggest a novel model in which rNMP-dependent hotspots are generated by sequential Top1 reactions and are consistent with rNMP-independent hotspots reflecting processing of a trapped Top1 cleavage complex.
Copyright © 2012 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

