A pilot study of rapid whole-genome sequencing for the investigation of a Legionella outbreak
- PMID: 23306006
- PMCID: PMC3553392
- DOI: 10.1136/bmjopen-2012-002175
A pilot study of rapid whole-genome sequencing for the investigation of a Legionella outbreak
Abstract
Objectives: Epidemiological investigations of Legionnaires' disease outbreaks rely on the rapid identification and typing of clinical and environmental Legionella isolates in order to identify and control the source of infection. Rapid bacterial whole-genome sequencing (WGS) is an emerging technology that has the potential to rapidly discriminate outbreak from non-outbreak isolates in a clinically relevant time frame.
Methods: We performed a pilot study to determine the feasibility of using bacterial WGS to differentiate outbreak from non-outbreak isolates collected during an outbreak of Legionnaires' disease. Seven Legionella isolates (three clinical and four environmental) were obtained from the reference laboratory and sequenced using the Illumina MiSeq platform at Addenbrooke's Hospital, Cambridge. Bioinformatic analysis was performed blinded to the epidemiological data at the Wellcome Trust Sanger Institute.
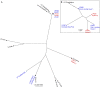
Results: We were able to distinguish outbreak from non-outbreak isolates using bacterial WGS, and to confirm the probable environmental source. Our analysis also highlighted constraints, which were the small number of Legionella pneumophila isolates available for sequencing, and the limited number of published genomes for comparison.
Conclusions: We have demonstrated the feasibility of using rapid WGS to investigate an outbreak of Legionnaires' disease. Future work includes building larger genomic databases of L pneumophila from both clinical and environmental sources, developing automated data interpretation software, and conducting a cost-benefit analysis of WGS versus current typing methods.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
