Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans
- PMID: 23306673
- PMCID: PMC3709822
- DOI: 10.1099/mic.0.064097-0
Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans
Abstract
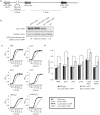
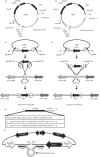
Candida albicans is the most prevalent fungal pathogen of humans. The current techniques used to construct C. albicans strains require integration of exogenous DNA at ectopic locations, which can exert position effects on gene expression that can confound the interpretation of data from critical experiments such as virulence assays. We have identified a large intergenic region, NEUT5L, which facilitates the integration and expression of ectopic genes. To construct and integrate inserts into this novel locus, we re-engineered yeast/bacterial shuttle vectors by incorporating 550 bp of homology to NEUT5L. These vectors allow rapid, facile cloning through in vivo recombination (gap repair) in Saccharomyces cerevisiae and efficient integration of the construct into the NEUT5L locus. Other useful features of these vectors include a choice of three selectable markers (URA3, the recyclable URA3-dpl200 or NAT1), and rare restriction enzyme recognition sites for releasing the insert from the vector prior to transformation into C. albicans, thereby reducing the insert size and preventing integration of non-C. albicans DNA. Importantly, unlike the commonly used RPS1/RP10 locus, integration at NEUT5L has no negative effect on growth rates and allows native-locus expression levels, making it an ideal genomic locus for the integration of exogenous DNA in C. albicans.
Figures

References
-
- Adams A., Gotschling D., Kaiser C., Stearns T. (1997). Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

