Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease
- PMID: 23307060
- PMCID: PMC3637979
- DOI: 10.1038/modpathol.2012.214
Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease
Abstract
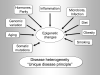
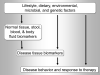
Epigenetics acts as an interface between environmental/exogenous factors, cellular responses, and pathological processes. Aberrant epigenetic signatures are a hallmark of complex multifactorial diseases (including neoplasms and malignancies such as leukemias, lymphomas, sarcomas, and breast, lung, prostate, liver, and colorectal cancers). Epigenetic signatures (DNA methylation, mRNA and microRNA expression, etc) may serve as biomarkers for risk stratification, early detection, and disease classification, as well as targets for therapy and chemoprevention. In particular, DNA methylation assays are widely applied to formalin-fixed, paraffin-embedded archival tissue specimens as clinical pathology tests. To better understand the interplay between etiological factors, cellular molecular characteristics, and disease evolution, the field of 'molecular pathological epidemiology (MPE)' has emerged as an interdisciplinary integration of 'molecular pathology' and 'epidemiology'. In contrast to traditional epidemiological research including genome-wide association studies (GWAS), MPE is founded on the unique disease principle, that is, each disease process results from unique profiles of exposomes, epigenomes, transcriptomes, proteomes, metabolomes, microbiomes, and interactomes in relation to the macroenvironment and tissue microenvironment. MPE may represent a logical evolution of GWAS, termed 'GWAS-MPE approach'. Although epigenome-wide association study attracts increasing attention, currently, it has a fundamental problem in that each cell within one individual has a unique, time-varying epigenome. Having a similar conceptual framework to systems biology, the holistic MPE approach enables us to link potential etiological factors to specific molecular pathology, and gain novel pathogenic insights on causality. The widespread application of epigenome (eg, methylome) analyses will enhance our understanding of disease heterogeneity, epigenotypes (CpG island methylator phenotype, LINE-1 (long interspersed nucleotide element-1; also called long interspersed nuclear element-1; long interspersed element-1; L1) hypomethylation, etc), and host-disease interactions. In this article, we illustrate increasing contribution of modern pathology to broader public health sciences, which attests pivotal roles of pathologists in the new integrated MPE science towards our ultimate goal of personalized medicine and prevention.
Figures

References
Publication types
MeSH terms
Grants and funding
- P01 CA087969/CA/NCI NIH HHS/United States
- R01 CA151993/CA/NCI NIH HHS/United States
- 1UM1 CA167552/CA/NCI NIH HHS/United States
- P01 CA87969/CA/NCI NIH HHS/United States
- P50 CA127003/CA/NCI NIH HHS/United States
- CAF/10/15/CSO_/Chief Scientist Office/United Kingdom
- R01 CA149222/CA/NCI NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R01 CA124908/CA/NCI NIH HHS/United States
- R01 CA137178/CA/NCI NIH HHS/United States
- R01 CA136950/CA/NCI NIH HHS/United States
- UM1 CA167552/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

