Rbm20 regulates titin alternative splicing as a splicing repressor
- PMID: 23307558
- PMCID: PMC3575840
- DOI: 10.1093/nar/gks1362
Rbm20 regulates titin alternative splicing as a splicing repressor
Abstract
Titin, a sarcomeric protein expressed primarily in striated muscles, is responsible for maintaining the structure and biomechanical properties of muscle cells. Cardiac titin undergoes developmental size reduction from 3.7 megadaltons in neonates to primarily 2.97 megadaltons in the adult. This size reduction results from gradually increased exon skipping between exons 50 and 219 of titin mRNA. Our previous study reported that Rbm20 is the splicing factor responsible for this process. In this work, we investigated its molecular mechanism. We demonstrate that Rbm20 mediates exon skipping by binding to titin pre-mRNA to repress the splicing of some regions; the exons/introns in these Rbm20-repressed regions are ultimately skipped. Rbm20 was also found to mediate intron retention and exon shuffling. The two Rbm20 speckles found in nuclei from muscle tissues were identified as aggregates of Rbm20 protein on the partially processed titin pre-mRNAs. Cooperative repression and alternative 3' splice site selection were found to be used by Rbm20 to skip different subsets of titin exons, and the splicing pathway selected depended on the ratio of Rbm20 to other splicing factors that vary with tissue type and developmental age.
Figures


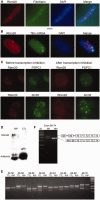

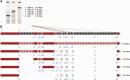


References
-
- Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 2001;89:1065–1072. - PubMed
-
- Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. - PubMed
-
- Maruyama K, Kimura S, Yoshidomi H, Sawada H, Kikuchi M. Molecular size and shape of beta-connection, an elastic protein of striated muscle. J. Biochem. 1984;95:1423–1433. - PubMed
-
- Trinick J, Knight P, Whiting A. Purification and properties of native titin. J. Mol. Biol. 1984;180:331–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

