Autophagy-related gene 12 (ATG12) is a novel determinant of primary resistance to HER2-targeted therapies: utility of transcriptome analysis of the autophagy interactome to guide breast cancer treatment
- PMID: 23307622
- PMCID: PMC3681498
- DOI: 10.18632/oncotarget.742
Autophagy-related gene 12 (ATG12) is a novel determinant of primary resistance to HER2-targeted therapies: utility of transcriptome analysis of the autophagy interactome to guide breast cancer treatment
Abstract
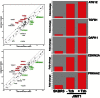
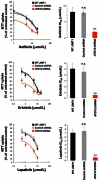
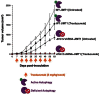
The autophagic process, which can facilitate breast cancer resistance to endocrine, cytotoxic, and molecularly targeted agents, is mainly regulated at the post-translational level. Although recent studies have suggested a possible transcriptome regulation of the autophagic genes, little is known about either the analysis tools that can be applied or the functional importance of putative candidate genes emerging from autophagy-dedicated transcriptome studies. In this context, we evaluated whether the constitutive activation of the autophagy machinery, as revealed by a transcriptome analysis using an autophagy-focused polymerase chain reaction (PCR) array, might allow for the identification of novel autophagy-specific biomarkers for intrinsic (primary) resistance to HER2-targeted therapies. Quantitative real-time PCR (qRT-PCR)-based profiling of 84 genes involved in autophagy revealed that, when compared to trastuzumab-sensitive SKBR3 cells, the positive regulator of autophagic vesicle formation ATG12 (autophagy-related gene 12) was the most differentially up-regulated gene in JIMT1 cells, a model of intrinsic cross-resistance to trastuzumab and other HER1/2-targeting drugs. An analysis of the transcriptional status of ATG12 in > 50 breast cancer cell lines suggested that the ATG12 transcript is commonly upregulated in trastuzumab-unresponsive HER2-overexpressing breast cancer cells. A lentiviral-delivered small hairpin RNA stable knockdown of the ATG12 gene fully suppressed the refractoriness of JIMT1 cells to trastuzumab, erlotinib, gefitinib, and lapatinib in vitro. ATG12 silencing significantly reduced JIMT1 tumor growth induced by subcutaneous injection in nude mice. Remarkably, the outgrowth of trastuzumab-unresponsive tumors was prevented completely when trastuzumab treatment was administered in an ATG12-silenced genetic background. We demonstrate for the first time the usefulness of low-density, autophagy-dedicated qRT-PCR-based platforms for monitoring primary resistance to HER2-targeted therapies by transcriptionally screening the autophagy interactome. The degree of predictive accuracy warrants further investigation in the clinical situation.
Conflict of interest statement
The authors of this manuscript have no conflicts of interest to declare.
Figures


Similar articles
-
HER2-positive breast cancer cells resistant to trastuzumab and lapatinib lose reliance upon HER2 and are sensitive to the multitargeted kinase inhibitor sorafenib.Breast Cancer Res Treat. 2011 Nov;130(1):29-40. doi: 10.1007/s10549-010-1281-5. Epub 2010 Dec 9. Breast Cancer Res Treat. 2011. PMID: 21153051
-
Different mechanisms for resistance to trastuzumab versus lapatinib in HER2-positive breast cancers--role of estrogen receptor and HER2 reactivation.Breast Cancer Res. 2011;13(6):R121. doi: 10.1186/bcr3067. Epub 2011 Nov 28. Breast Cancer Res. 2011. PMID: 22123186 Free PMC article.
-
Upregulation of mucin4 in ER-positive/HER2-overexpressing breast cancer xenografts with acquired resistance to endocrine and HER2-targeted therapies.Breast Cancer Res Treat. 2012 Jul;134(2):583-93. doi: 10.1007/s10549-012-2082-9. Epub 2012 May 29. Breast Cancer Res Treat. 2012. PMID: 22644656 Free PMC article.
-
Beyond trastuzumab and lapatinib: new options for HER2-positive breast cancer.Am Soc Clin Oncol Educ Book. 2013. doi: 10.14694/EdBook_AM.2013.33.e2. Am Soc Clin Oncol Educ Book. 2013. PMID: 23714441 Review.
-
Current and future anti-HER2 therapy in breast cancer.J BUON. 2013 Jan-Mar;18(1):4-16. J BUON. 2013. PMID: 23613383 Review.
Cited by
-
Acquired resistance to metformin in breast cancer cells triggers transcriptome reprogramming toward a degradome-related metastatic stem-like profile.Cell Cycle. 2014;13(7):1132-44. doi: 10.4161/cc.27982. Epub 2014 Feb 7. Cell Cycle. 2014. PMID: 24553122 Free PMC article.
-
Autophagy and Apoptotic Crosstalk: Mechanism of Therapeutic Resistance in HER2-Positive Breast Cancer.Breast Cancer (Auckl). 2016 Mar 13;10:13-23. doi: 10.4137/BCBCR.S32791. eCollection 2016. Breast Cancer (Auckl). 2016. PMID: 26997868 Free PMC article. Review.
-
The autophagy inhibitor verteporfin moderately enhances the antitumor activity of gemcitabine in a pancreatic ductal adenocarcinoma model.J Cancer. 2013 Aug 28;4(7):585-96. doi: 10.7150/jca.7030. eCollection 2013. J Cancer. 2013. PMID: 24069069 Free PMC article.
-
Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer.Mol Cancer. 2017 Nov 21;16(1):174. doi: 10.1186/s12943-017-0743-3. Mol Cancer. 2017. PMID: 29162158 Free PMC article.
-
miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2.Cell Death Dis. 2015 May 21;6(5):e1766. doi: 10.1038/cddis.2015.123. Cell Death Dis. 2015. PMID: 25996293 Free PMC article.
References
-
- Nahta R, Yu D, Hung MC, Hortobagyi GN, Esteva FJ. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat Clin Pract Oncol. 2006;3:269–280. - PubMed
-
- Chen FL, Xia W, Spector NL. Acquired resistance to small molecule ErbB2 tyrosine kinase inhibitors. Clin Cancer Res. 2008;14:6730–6734. - PubMed
-
- Spector NL, Blackwell KL. Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol. 2009;27:5838–5847. - PubMed
-
- Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2011;9:16–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

