Regulation of AID, the B-cell genome mutator
- PMID: 23307864
- PMCID: PMC3553278
- DOI: 10.1101/gad.200014.112
Regulation of AID, the B-cell genome mutator
Abstract
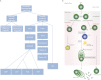
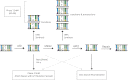
The mechanisms by which B cells somatically engineer their genomes to generate the vast diversity of antibodies required to challenge the nearly infinite number of antigens that immune systems encounter are of tremendous clinical and academic interest. The DNA cytidine deaminase activation-induced deaminase (AID) catalyzes two of these mechanisms: class switch recombination (CSR) and somatic hypermutation (SHM). Recent discoveries indicate a significant promiscuous targeting of this B-cell mutator enzyme genome-wide. Here we discuss the various regulatory elements that control AID activity and prevent AID from inducing genomic instability and thereby initiating oncogenesis.
Figures




References
-
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell 17: 103–112 - PubMed
-
- Allen CD, Okada T, Tang HL, Cyster JG 2007b. Imaging of germinal center selection events during affinity maturation. Science 315: 528–531 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
