Gene copy-number variation in haploid and diploid strains of the yeast Saccharomyces cerevisiae
- PMID: 23307895
- PMCID: PMC3583998
- DOI: 10.1534/genetics.112.146522
Gene copy-number variation in haploid and diploid strains of the yeast Saccharomyces cerevisiae
Abstract
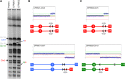
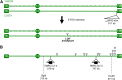
The increasing ability to sequence and compare multiple individual genomes within a species has highlighted the fact that copy-number variation (CNV) is a substantial and underappreciated source of genetic diversity. Chromosome-scale mutations occur at rates orders of magnitude higher than base substitutions, yet our understanding of the mechanisms leading to CNVs has been lagging. We examined CNV in a region of chromosome 5 (chr5) in haploid and diploid strains of Saccharomyces cerevisiae. We optimized a CNV detection assay based on a reporter cassette containing the SFA1 and CUP1 genes that confer gene dosage-dependent tolerance to formaldehyde and copper, respectively. This optimized reporter allowed the selection of low-order gene amplification events, going from one copy to two copies in haploids and from two to three copies in diploids. In haploid strains, most events involved tandem segmental duplications mediated by nonallelic homologous recombination between flanking direct repeats, primarily Ty1 elements. In diploids, most events involved the formation of a recurrent nonreciprocal translocation between a chr5 Ty1 element and another Ty1 repeat on chr13. In addition to amplification events, a subset of clones displaying elevated resistance to formaldehyde had point mutations within the SFA1 coding sequence. These mutations were all dominant and are proposed to result in hyperactive forms of the formaldehyde dehydrogenase enzyme.
Figures




References
-
- Brown C. J., Todd K. M., Rosenzweig R. F., 1998. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol. Biol. Evol. 15: 931–942. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

