c-Myc suppression of DNA double-strand break repair
- PMID: 23308051
- PMCID: PMC3540944
- DOI: 10.1593/neo.121258
c-Myc suppression of DNA double-strand break repair
Abstract
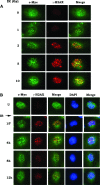
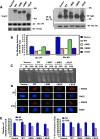
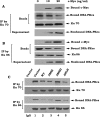
c-Myc is a transcriptional factor that functions as a central regulator of cell growth, proliferation, and apoptosis. Overexpression of c-Myc also enhances DNA double-strand breaks (DSBs), genetic instability, and tumorigenesis. However, the mechanism(s) involved remains elusive. Here, we discovered that γ-ray ionizing radiation-induced DSBs promote c-Myc to form foci and to co-localize with γ-H2AX. Conditional expression of c-Myc in HO15.19 c-Myc null cells using the Tet-Off/Tet-On inducible system results in down-regulation of Ku DNA binding and suppressed activities of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and DNA end-joining, leading to inhibition of DSB repair and enhanced chromosomal and chromatid breaks. Expression of c-Myc reduces both signal and coding joins with decreased fidelity during V(D)J recombination. Mechanistically, c-Myc directly interacts with Ku70 protein through its Myc box II (MBII) domain. Removal of the MBII domain from c-Myc abrogates its inhibitory effects on Ku DNA binding, DNA-PKcs, and DNA end-joining activities, which results in loss of c-Myc's ability to block DSB repair and V(D)J recombination. Interestingly, c-Myc directly disrupts the Ku/DNA-PKcs complex in vitro and in vivo. Thus, c-Myc suppression of DSB repair and V(D)J recombination may occur through inhibition of the nonhomologous end-joining pathway, which provides insight into the mechanism of c-Myc in the development of tumors through promotion of genomic instability.
Figures



Similar articles
-
Functional redundancy between the XLF and DNA-PKcs DNA repair factors in V(D)J recombination and nonhomologous DNA end joining.Proc Natl Acad Sci U S A. 2013 Feb 5;110(6):2234-9. doi: 10.1073/pnas.1222573110. Epub 2013 Jan 23. Proc Natl Acad Sci U S A. 2013. PMID: 23345432 Free PMC article.
-
A nonhomologous end-joining pathway is required for protein phosphatase 2A promotion of DNA double-strand break repair.Neoplasia. 2009 Oct;11(10):1012-21. doi: 10.1593/neo.09720. Neoplasia. 2009. PMID: 19794960 Free PMC article.
-
Bcl2 negatively regulates DNA double-strand-break repair through a nonhomologous end-joining pathway.Mol Cell. 2008 Feb 29;29(4):488-98. doi: 10.1016/j.molcel.2007.12.029. Mol Cell. 2008. PMID: 18313386 Free PMC article.
-
The Ku heterodimer: function in DNA repair and beyond.Mutat Res Rev Mutat Res. 2015 Jan-Mar;763:15-29. doi: 10.1016/j.mrrev.2014.06.002. Epub 2014 Jul 4. Mutat Res Rev Mutat Res. 2015. PMID: 25795113 Review.
-
Ku, a DNA repair protein with multiple cellular functions?Mutat Res. 1999 May 14;434(1):3-15. doi: 10.1016/s0921-8777(99)00006-3. Mutat Res. 1999. PMID: 10377944 Review.
Cited by
-
Role of Ku70 in deubiquitination of Mcl-1 and suppression of apoptosis.Cell Death Differ. 2014 Jul;21(7):1160-9. doi: 10.1038/cdd.2014.42. Epub 2014 Apr 25. Cell Death Differ. 2014. PMID: 24769731 Free PMC article.
-
Identification of synthetic lethality of PRKDC in MYC-dependent human cancers by pooled shRNA screening.BMC Cancer. 2014 Dec 13;14:944. doi: 10.1186/1471-2407-14-944. BMC Cancer. 2014. PMID: 25495526 Free PMC article.
-
Effects of Low-Dose Bisphenol A on DNA Damage and Proliferation of Breast Cells: The Role of c-Myc.Environ Health Perspect. 2015 Dec;123(12):1271-9. doi: 10.1289/ehp.1409199. Epub 2015 May 1. Environ Health Perspect. 2015. PMID: 25933419 Free PMC article.
-
Synergistic Effect of Ubiquitin-Specific Protease 14 and Poly(ADP-Ribose) Glycohydrolase Co-Inhibition in BRCA1-Mutant, Poly(ADP-Ribose) Polymerase Inhibitor-Resistant Triple-Negative Breast Cancer Cells.Onco Targets Ther. 2024 Sep 6;17:741-753. doi: 10.2147/OTT.S463217. eCollection 2024. Onco Targets Ther. 2024. PMID: 39258222 Free PMC article.
-
c-MYC-induced genomic instability.Cold Spring Harb Perspect Med. 2014 Apr 1;4(4):a014373. doi: 10.1101/cshperspect.a014373. Cold Spring Harb Perspect Med. 2014. PMID: 24692190 Free PMC article. Review.
References
-
- Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. - PubMed
-
- Rothkamm K, Kuhne M, Jeggo PA, Lobrich M. Radiation-induced genomic rearrangements formed by nonhomologous end-joining of DNA double-strand breaks. Cancer Res. 2001;61:3886–3893. - PubMed
-
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. - PubMed
Supplemental References
-
- Shin KH, Kang MK, Dicterow E, Kameta A, Baluda MA, Park NH. Introduction of human telomerase reverse transcriptase to normal human fibroblasts enhances DNA repair capacity. Clin Cancer Res. 2004;10:2551–2560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
