Virus-encoded microRNAs: an overview and a look to the future
- PMID: 23308061
- PMCID: PMC3534370
- DOI: 10.1371/journal.ppat.1003018
Virus-encoded microRNAs: an overview and a look to the future
Abstract
MicroRNAs (miRNAs) are small RNAs that play important roles in the regulation of gene expression. First described as posttranscriptional gene regulators in eukaryotic hosts, virus-encoded miRNAs were later uncovered. It is now apparent that diverse virus families, most with DNA genomes, but at least some with RNA genomes, encode miRNAs. While deciphering the functions of viral miRNAs has lagged behind their discovery, recent functional studies are bringing into focus these roles. Some of the best characterized viral miRNA functions include subtle roles in prolonging the longevity of infected cells, evading the immune response, and regulating the switch to lytic infection. Notably, all of these functions are particularly important during persistent infections. Furthermore, an emerging view of viral miRNAs suggests two distinct groups exist. In the first group, viral miRNAs mimic host miRNAs and take advantage of conserved networks of host miRNA target sites. In the larger second group, viral miRNAs do not share common target sites conserved for host miRNAs, and it remains unclear what fraction of these targeted transcripts are beneficial to the virus. Recent insights from multiple virus families have revealed new ways of interacting with the host miRNA machinery including noncanonical miRNA biogenesis and new mechanisms of posttranscriptional cis gene regulation. Exciting challenges await the field, including determining the most relevant miRNA targets and parlaying our current understanding of viral miRNAs into new therapeutic strategies. To accomplish these goals and to better grasp miRNA function, new in vivo models that recapitulate persistent infections associated with viral pathogens are required.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

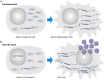
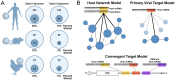

References
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. - PubMed
-
- Buchon N, Vaury C (2006) RNAi: a defensive RNA-silencing against viruses and transposable elements. Heredity 96: 195–202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

