Transmission of equine influenza virus during an outbreak is characterized by frequent mixed infections and loose transmission bottlenecks
- PMID: 23308065
- PMCID: PMC3534375
- DOI: 10.1371/journal.ppat.1003081
Transmission of equine influenza virus during an outbreak is characterized by frequent mixed infections and loose transmission bottlenecks
Abstract
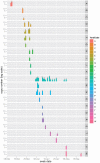
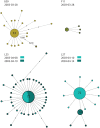
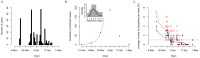
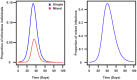
The ability of influenza A viruses (IAVs) to cross species barriers and evade host immunity is a major public health concern. Studies on the phylodynamics of IAVs across different scales - from the individual to the population - are essential for devising effective measures to predict, prevent or contain influenza emergence. Understanding how IAVs spread and evolve during outbreaks is critical for the management of epidemics. Reconstructing the transmission network during a single outbreak by sampling viral genetic data in time and space can generate insights about these processes. Here, we obtained intra-host viral sequence data from horses infected with equine influenza virus (EIV) to reconstruct the spread of EIV during a large outbreak. To this end, we analyzed within-host viral populations from sequences covering 90% of the infected yards. By combining gene sequence analyses with epidemiological data, we inferred a plausible transmission network, in turn enabling the comparison of transmission patterns during the course of the outbreak and revealing important epidemiological features that were not apparent using either approach alone. The EIV populations displayed high levels of genetic diversity, and in many cases we observed distinct viral populations containing a dominant variant and a number of related minor variants that were transmitted between infectious horses. In addition, we found evidence of frequent mixed infections and loose transmission bottlenecks in these naturally occurring populations. These frequent mixed infections likely influence the size of epidemics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, et al. (2009) Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459: 1122–U1107. - PubMed
-
- Grenfell BT, Pybus OG, Gog JR, Wood JL, Daly JM, et al. (2004) Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303: 327–332. - PubMed
-
- Webster RG (1993) Are equine 1 influenza viruses still present in horses? Equine Vet J 25: 537–538. - PubMed

