A unified model of the GABA(A) receptor comprising agonist and benzodiazepine binding sites
- PMID: 23308109
- PMCID: PMC3538749
- DOI: 10.1371/journal.pone.0052323
A unified model of the GABA(A) receptor comprising agonist and benzodiazepine binding sites
Abstract
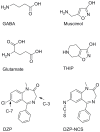
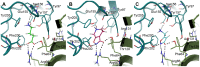
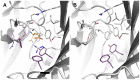
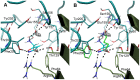
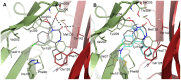
We present a full-length α(1)β(2)γ(2) GABA receptor model optimized for agonists and benzodiazepine (BZD) allosteric modulators. We propose binding hypotheses for the agonists GABA, muscimol and THIP and for the allosteric modulator diazepam (DZP). The receptor model is primarily based on the glutamate-gated chloride channel (GluCl) from C. elegans and includes additional structural information from the prokaryotic ligand-gated ion channel ELIC in a few regions. Available mutational data of the binding sites are well explained by the model and the proposed ligand binding poses. We suggest a GABA binding mode similar to the binding mode of glutamate in the GluCl X-ray structure. Key interactions are predicted with residues α(1)R66, β(2)T202, α(1)T129, β(2)E155, β(2)Y205 and the backbone of β(2)S156. Muscimol is predicted to bind similarly, however, with minor differences rationalized with quantum mechanical energy calculations. Muscimol key interactions are predicted to be α(1)R66, β(2)T202, α(1)T129, β(2)E155, β(2)Y205 and β(2)F200. Furthermore, we argue that a water molecule could mediate further interactions between muscimol and the backbone of β(2)S156 and β(2)Y157. DZP is predicted to bind with interactions comparable to those of the agonists in the orthosteric site. The carbonyl group of DZP is predicted to interact with two threonines α(1)T206 and γ(2)T142, similar to the acidic moiety of GABA. The chlorine atom of DZP is placed near the important α(1)H101 and the N-methyl group near α(1)Y159, α(1)T206, and α(1)Y209. We present a binding mode of DZP in which the pending phenyl moiety of DZP is buried in the binding pocket and thus shielded from solvent exposure. Our full length GABA(A) receptor is made available as Model S1.
Conflict of interest statement
Figures



References
-
- Johnston GA (2005) GABA(A) receptor channel pharmacology. Curr Pharm Des 11: 1867–1885. - PubMed
-
- Johnston GA, Curtis DR, De Groat WC, Duggan AW (1968) Central actions of ibotenic acid and muscimol. Biochem Pharmacol 17: 2488–2489. - PubMed
-
- Ebert B, Frolund B, Diemer NH, Krogsgaard-Larsen P (1999) Equilibrium binding characteristics of [3H]thiomuscimol. Neurochem Int 34: 427–434. - PubMed
-
- Krogsgaard-Larsen P, Johnston GA, Lodge D, Curtis DR (1977) A new class of GABA agonist. Nature 268: 53–55. - PubMed
-
- Frolund B, Kristiansen U, Brehm L, Hansen AB, Krogsgaard-Larsen P, et al. (1995) Partial GABAA receptor agonists. Synthesis and in vitro pharmacology of a series of nonannulated analogs of 4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridin-3-ol. J Med Chem 38: 3287–3296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

