Automatic peak selection by a Benjamini-Hochberg-based algorithm
- PMID: 23308147
- PMCID: PMC3538655
- DOI: 10.1371/journal.pone.0053112
Automatic peak selection by a Benjamini-Hochberg-based algorithm
Abstract
A common issue in bioinformatics is that computational methods often generate a large number of predictions sorted according to certain confidence scores. A key problem is then determining how many predictions must be selected to include most of the true predictions while maintaining reasonably high precision. In nuclear magnetic resonance (NMR)-based protein structure determination, for instance, computational peak picking methods are becoming more and more common, although expert-knowledge remains the method of choice to determine how many peaks among thousands of candidate peaks should be taken into consideration to capture the true peaks. Here, we propose a Benjamini-Hochberg (B-H)-based approach that automatically selects the number of peaks. We formulate the peak selection problem as a multiple testing problem. Given a candidate peak list sorted by either volumes or intensities, we first convert the peaks into [Formula: see text]-values and then apply the B-H-based algorithm to automatically select the number of peaks. The proposed approach is tested on the state-of-the-art peak picking methods, including WaVPeak [1] and PICKY [2]. Compared with the traditional fixed number-based approach, our approach returns significantly more true peaks. For instance, by combining WaVPeak or PICKY with the proposed method, the missing peak rates are on average reduced by 20% and 26%, respectively, in a benchmark set of 32 spectra extracted from eight proteins. The consensus of the B-H-selected peaks from both WaVPeak and PICKY achieves 88% recall and 83% precision, which significantly outperforms each individual method and the consensus method without using the B-H algorithm. The proposed method can be used as a standard procedure for any peak picking method and straightforwardly applied to some other prediction selection problems in bioinformatics. The source code, documentation and example data of the proposed method is available at http://sfb.kaust.edu.sa/pages/software.aspx.
Conflict of interest statement
Figures
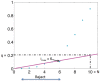
 ) is 10 and the false discovery proportion (
) is 10 and the false discovery proportion ( ) is 0.2. The largest index of the hypotheses that is below the line is 6 (
) is 0.2. The largest index of the hypotheses that is below the line is 6 ( ). Therefore, the first six hypotheses are rejected as the predicted peaks.
). Therefore, the first six hypotheses are rejected as the predicted peaks.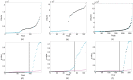
 and B-H procedure are shown in black and magenta, respectively.
and B-H procedure are shown in black and magenta, respectively.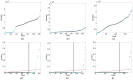
 and the B-H procedure are shown in black and magenta, respectively.
and the B-H procedure are shown in black and magenta, respectively.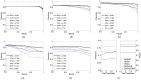
 consensus method; the solid cyan curves are for B-H WaVPeak; the dashed cyan curves are for the original WaVPeak; the solid magenta curves are for B-H PICKY; and the dashed magenta curves are for the original PICKY. The relative area under curve (AUC) values are in legends, which are the area under curve over the total area of recall at least 0.7. (f): sensitivity analysis for different number of peaks. The precision and recall values of B-H WaVPeak are shown when
consensus method; the solid cyan curves are for B-H WaVPeak; the dashed cyan curves are for the original WaVPeak; the solid magenta curves are for B-H PICKY; and the dashed magenta curves are for the original PICKY. The relative area under curve (AUC) values are in legends, which are the area under curve over the total area of recall at least 0.7. (f): sensitivity analysis for different number of peaks. The precision and recall values of B-H WaVPeak are shown when  ,
,  ,
,  and
and  top peaks are used to calculate the p-values.
top peaks are used to calculate the p-values.References
-
- Wüthrich K (1986) NMR of Proteins and Nucleic Acids. New York: John Wiley and Sons.
-
- Gao X (2009) Towards automating protein structure determination from NMR data. PhD dissertation, University of Waterloo.
-
- Gao X (2012) Mathematical approaches to the NMR peak-picking problem. Journal of Applied and Computational Mathematics 1: 1.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

