RNAP-II molecules participate in the anchoring of the ORC to rDNA replication origins
- PMID: 23308214
- PMCID: PMC3537633
- DOI: 10.1371/journal.pone.0053405
RNAP-II molecules participate in the anchoring of the ORC to rDNA replication origins
Abstract
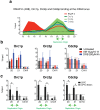
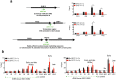
The replication of genomic DNA is limited to a single round per cell cycle. The first component, which recognises and remains bound to origins from recognition until activation and replication elongation, is the origin recognition complex. How origin recognition complex (ORC) proteins remain associated with chromatin throughout the cell cycle is not yet completely understood. Several genome-wide studies have undoubtedly demonstrated that RNA polymerase II (RNAP-II) binding sites overlap with replication origins and with the binding sites of the replication components. RNAP-II is no longer merely associated with transcription elongation. Several reports have demonstrated that RNAP-II molecules affect chromatin structure, transcription, mRNA processing, recombination and DNA repair, among others. Most of these activities have been reported to directly depend on the interaction of proteins with the C-terminal domain (CTD) of RNAP-II. Two-dimensional gels results and ChIP analysis presented herein suggest that stalled RNAP-II molecules bound to the rDNA chromatin participate in the anchoring of ORC proteins to origins during the G1 and S-phases. The results show that in the absence of RNAP-II, Orc1p, Orc2p and Cdc6p do not bind to origins. Moreover, co-immunoprecipitation experiments suggest that Ser2P-CTD and hypophosphorylated RNAP-II interact with Orc1p. In the context of rDNA, cryptic transcription by RNAP-II did not negatively interfere with DNA replication. However, the results indicate that RNAP-II is not necessary to maintain the binding of ORCs to the origins during metaphase. These findings highlight for the first time the potential importance of stalled RNAP-II in the regulation of DNA replication.
Conflict of interest statement
Figures





Similar articles
-
The budding yeast Fkh1 Forkhead associated (FHA) domain promotes a G1-chromatin state and the activity of chromosomal DNA replication origins.PLoS Genet. 2024 Aug 5;20(8):e1011366. doi: 10.1371/journal.pgen.1011366. eCollection 2024 Aug. PLoS Genet. 2024. PMID: 39102423 Free PMC article.
-
The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin.Genes Dev. 2010 Jul 1;24(13):1418-33. doi: 10.1101/gad.1906410. Genes Dev. 2010. PMID: 20595233 Free PMC article.
-
Yeast two-hybrid analysis of the origin recognition complex of Saccharomyces cerevisiae: interaction between subunits and identification of binding proteins.FEMS Yeast Res. 2007 Dec;7(8):1263-9. doi: 10.1111/j.1567-1364.2007.00298.x. Epub 2007 Sep 6. FEMS Yeast Res. 2007. PMID: 17825065
-
Dynamics of pre-replication complex proteins during the cell division cycle.Philos Trans R Soc Lond B Biol Sci. 2004 Jan 29;359(1441):7-16. doi: 10.1098/rstb.2003.1360. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15065651 Free PMC article. Review.
-
Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle.J Cell Sci Suppl. 1995;19:67-72. doi: 10.1242/jcs.1995.supplement_19.9. J Cell Sci Suppl. 1995. PMID: 8655649 Review.
Cited by
-
The role of transcription in the activation of a Drosophila amplification origin.G3 (Bethesda). 2014 Oct 14;4(12):2403-8. doi: 10.1534/g3.114.014050. G3 (Bethesda). 2014. PMID: 25320071 Free PMC article.
-
Noncoding transcription influences the replication initiation program through chromatin regulation.Genome Res. 2018 Dec;28(12):1882-1893. doi: 10.1101/gr.239582.118. Epub 2018 Nov 6. Genome Res. 2018. PMID: 30401734 Free PMC article.
-
RNA polymerase II C-terminal domain: Tethering transcription to transcript and template.Chem Rev. 2013 Nov 13;113(11):8423-55. doi: 10.1021/cr400158h. Epub 2013 Sep 16. Chem Rev. 2013. PMID: 24040939 Free PMC article. Review. No abstract available.
-
Endogenous single-strand DNA breaks at RNA polymerase II promoters in Saccharomyces cerevisiae.Nucleic Acids Res. 2018 Nov 16;46(20):10649-10668. doi: 10.1093/nar/gky743. Nucleic Acids Res. 2018. PMID: 30445637 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

