The NifA-RpoN regulon of Mesorhizobium loti strain R7A and its symbiotic activation by a novel LacI/GalR-family regulator
- PMID: 23308282
- PMCID: PMC3538637
- DOI: 10.1371/journal.pone.0053762
The NifA-RpoN regulon of Mesorhizobium loti strain R7A and its symbiotic activation by a novel LacI/GalR-family regulator
Abstract
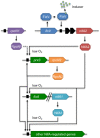
Mesorhizobium loti is the microsymbiont of Lotus species, including the model legume L. japonicus. M. loti differs from other rhizobia in that it contains two copies of the key nitrogen fixation regulatory gene nifA, nifA1 and nifA2, both of which are located on the symbiosis island ICEMlSym(R7A). M. loti R7A also contains two rpoN genes, rpoN1 located on the chromosome outside of ICEMlSym(R7A) and rpoN2 that is located on ICEMlSym(R7A). The aims of the current work were to establish how nifA expression was activated in M. loti and to characterise the NifA-RpoN regulon. The nifA2 and rpoN2 genes were essential for nitrogen fixation whereas nifA1 and rpoN1 were dispensable. Expression of nifA2 was activated, possibly in response to an inositol derivative, by a novel regulator of the LacI/GalR family encoded by the fixV gene located upstream of nifA2. Other than the well-characterized nif/fix genes, most NifA2-regulated genes were not required for nitrogen fixation although they were strongly expressed in nodules. The NifA-regulated nifZ and fixU genes, along with nifQ which was not NifA-regulated, were required in M. loti for a fully effective symbiosis although they are not present in some other rhizobia. The NifA-regulated gene msi158 that encodes a porin was also required for a fully effective symbiosis. Several metabolic genes that lacked NifA-regulated promoters were strongly expressed in nodules in a NifA2-dependent manner but again mutants did not have an overt symbiotic phenotype. In summary, many genes encoded on ICEMlSym(R7A) were strongly expressed in nodules but not free-living rhizobia, but were not essential for symbiotic nitrogen fixation. It seems likely that some of these genes have functional homologues elsewhere in the genome and that bacteroid metabolism may be sufficiently plastic to adapt to loss of certain enzymatic functions.
Conflict of interest statement
Figures


References
-
- Ramsay JP, Sullivan JT, Stuart GS, Lamont IL, Ronson CW (2006) Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol Microbiol 62: 723–734. - PubMed
-
- Wozniak RA, Waldor MK (2010) Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8: 552–563. - PubMed
-
- Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, et al. (2000) Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti . DNA Res 7: 331–338. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

