The utility of chromosomal microarray analysis in developmental and behavioral pediatrics
- PMID: 23311723
- PMCID: PMC3725967
- DOI: 10.1111/cdev.12050
The utility of chromosomal microarray analysis in developmental and behavioral pediatrics
Abstract
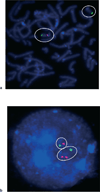
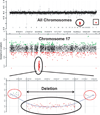
Chromosomal microarray analysis (CMA) has emerged as a powerful new tool to identify genomic abnormalities associated with a wide range of developmental disabilities including congenital malformations, cognitive impairment, and behavioral abnormalities. CMA includes array comparative genomic hybridization (CGH) and single nucleotide polymorphism (SNP) arrays, both of which are useful for detection of genomic copy number variants (CNV) such as microdeletions and microduplications. The frequency of disease-causing CNVs is highest (20%-25%) in children with moderate to severe intellectual disability accompanied by malformations or dysmorphic features. Disease-causing CNVs are found in 5%-10% of cases of autism, being more frequent in severe phenotypes. CMA has replaced Giemsa-banded karyotype as the first-tier test for genetic evaluation of children with developmental and behavioral disabilities.
© 2013 The Author. Child Development © 2013 Society for Research in Child Development, Inc.
Conflict of interest statement
Conflict of interest: The author is Professor and Chair of the Department of Molecular and Human Genetics at Baylor College of Medicine (BCM); the Department offers extensive genetic laboratory testing and derives revenue from this activity.
Figures


Similar articles
-
Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features.Eur J Paediatr Neurol. 2013 Nov;17(6):589-99. doi: 10.1016/j.ejpn.2013.04.010. Epub 2013 May 24. Eur J Paediatr Neurol. 2013. PMID: 23711909
-
[The diagnostic value of chromosome microarray analysis technique in the genetic causes of children with intellectual disability or global developmental delay].Zhonghua Yi Xue Za Zhi. 2021 Jan 19;101(3):224-228. doi: 10.3760/cma.j.cn112137-20200422-01275. Zhonghua Yi Xue Za Zhi. 2021. PMID: 33455150 Chinese.
-
The clinical benefit of array-based comparative genomic hybridization for detection of copy number variants in Czech children with intellectual disability and developmental delay.BMC Med Genomics. 2019 Jul 23;12(1):111. doi: 10.1186/s12920-019-0559-7. BMC Med Genomics. 2019. PMID: 31337399 Free PMC article.
-
Genomic copy number variation in disorders of cognitive development.J Am Acad Child Adolesc Psychiatry. 2010 Nov;49(11):1091-104. doi: 10.1016/j.jaac.2010.08.009. J Am Acad Child Adolesc Psychiatry. 2010. PMID: 20970697 Free PMC article. Review.
-
Experience of chromosomal microarray applied in prenatal and postnatal settings in Hong Kong.Am J Med Genet C Semin Med Genet. 2019 Jun;181(2):196-207. doi: 10.1002/ajmg.c.31697. Epub 2019 Mar 23. Am J Med Genet C Semin Med Genet. 2019. PMID: 30903683 Review.
Cited by
-
Supporting interoperability of genetic data with LOINC.J Am Med Inform Assoc. 2015 May;22(3):621-7. doi: 10.1093/jamia/ocu012. Epub 2015 Feb 5. J Am Med Inform Assoc. 2015. PMID: 25656513 Free PMC article.
-
Parents' perceptions of the usefulness of chromosomal microarray analysis for children with autism spectrum disorders.J Autism Dev Disord. 2015 Oct;45(10):3262-75. doi: 10.1007/s10803-015-2489-3. J Autism Dev Disord. 2015. PMID: 26066358 Free PMC article.
-
Chromosomal microarray analysis in clinical evaluation of neurodevelopmental disorders-reporting a novel deletion of SETDB1 and illustration of counseling challenge.Pediatr Res. 2016 Sep;80(3):371-81. doi: 10.1038/pr.2016.101. Epub 2016 Apr 27. Pediatr Res. 2016. PMID: 27119313 Free PMC article.
-
Diagnostic and Therapeutic Misconception: Parental Expectations and Perspectives Regarding Genetic Testing for Developmental Disorders.J Autism Dev Disord. 2019 Jan;49(1):363-375. doi: 10.1007/s10803-018-3768-6. J Autism Dev Disord. 2019. PMID: 30284667
-
A New Patient with Potocki-Lupski Syndrome: A Literature Review.J Pediatr Genet. 2018 Mar;7(1):29-34. doi: 10.1055/s-0037-1604479. Epub 2017 Jul 27. J Pediatr Genet. 2018. PMID: 29441219 Free PMC article.
References
-
- American College of Obstetricians and Gynecologists. Invasive prenatal testing for aneuploidy. Obstetrics and Gynecology. 2007;110:1459–1467. ACOG practice bulletin no. 88, December 2007. - PubMed
-
- Astrom RL, Wadsworth SJ, DeFries JC. Etiology of the stability of reading difficulties: The longitudinal twin study of reading disabilities. Twin Research and Human Genetics. 2007;10:434–439. - PubMed
-
- Ballif BC, Sulpizio SG, Lloyd RM, Minier SL, Theisen A, Bejjani BA, et al. The clinical utility of enhanced subtelomeric coverage in array CGH. American Journal of Medical Genetics A. 2007;143A:1850–1857. - PubMed
-
- Berg JS, Brunetti-Pierri N, Peters SU, Kang SH, Fong CT, Salamone J, et al. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams-Beuren syndrome region. Genetic Medicine. 2007;9:427–441. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

