mTOR direct interactions with Rheb-GTPase and raptor: sub-cellular localization using fluorescence lifetime imaging
- PMID: 23311891
- PMCID: PMC3549280
- DOI: 10.1186/1471-2121-14-3
mTOR direct interactions with Rheb-GTPase and raptor: sub-cellular localization using fluorescence lifetime imaging
Abstract
Background: The mammalian target of rapamycin (mTOR) signalling pathway has a key role in cellular regulation and several diseases. While it is thought that Rheb GTPase regulates mTOR, acting immediately upstream, while raptor is immediately downstream of mTOR, direct interactions have yet to be verified in living cells, furthermore the localisation of Rheb has been reported to have only a cytoplasmic cellular localization.
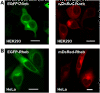
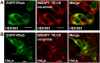
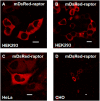
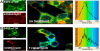
Results: In this study a cytoplasmic as well as a significant sub-cellular nuclear mTOR localization was shown , utilizing green and red fluorescent protein (GFP and DsRed) fusion and highly sensitive single photon counting fluorescence lifetime imaging microscopy (FLIM) of live cells. The interaction of the mTORC1 components Rheb, mTOR and raptor, tagged with EGFP/DsRed was determined using fluorescence energy transfer-FLIM. The excited-state lifetime of EGFP-mTOR of ~2400 ps was reduced by energy transfer to ~2200 ps in the cytoplasm and to 2000 ps in the nucleus when co-expressed with DsRed-Rheb, similar results being obtained for co-expressed EGFP-mTOR and DsRed-raptor. The localization and distribution of mTOR was modified by amino acid withdrawal and re-addition but not by rapamycin.
Conclusions: The results illustrate the power of GFP-technology combined with FRET-FLIM imaging in the study of the interaction of signalling components in living cells, here providing evidence for a direct physical interaction between mTOR and Rheb and between mTOR and raptor in living cells for the first time.
Figures









Similar articles
-
Raptor and Rheb negatively regulate skeletal myogenesis through suppression of insulin receptor substrate 1 (IRS1).J Biol Chem. 2011 Oct 14;286(41):35675-35682. doi: 10.1074/jbc.M111.262881. Epub 2011 Aug 18. J Biol Chem. 2011. PMID: 21852229 Free PMC article.
-
Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein.J Biol Chem. 2009 May 8;284(19):12783-91. doi: 10.1074/jbc.M809207200. Epub 2009 Mar 19. J Biol Chem. 2009. PMID: 19299511 Free PMC article.
-
Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling.Oncogene. 2010 Jan 21;29(3):380-91. doi: 10.1038/onc.2009.336. Epub 2009 Oct 19. Oncogene. 2010. PMID: 19838215 Free PMC article.
-
Amino acid regulation of TOR complex 1.Am J Physiol Endocrinol Metab. 2009 Apr;296(4):E592-602. doi: 10.1152/ajpendo.90645.2008. Epub 2008 Sep 2. Am J Physiol Endocrinol Metab. 2009. PMID: 18765678 Free PMC article. Review.
-
Rag GTPase in amino acid signaling.Amino Acids. 2016 Apr;48(4):915-928. doi: 10.1007/s00726-016-2171-x. Epub 2016 Jan 18. Amino Acids. 2016. PMID: 26781224 Review.
Cited by
-
Roles of mTOR complexes in the kidney: implications for renal disease and transplantation.Nat Rev Nephrol. 2016 Oct;12(10):587-609. doi: 10.1038/nrneph.2016.108. Epub 2016 Aug 1. Nat Rev Nephrol. 2016. PMID: 27477490 Free PMC article. Review.
-
Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation.Sci Rep. 2016 Aug 22;6:31697. doi: 10.1038/srep31697. Sci Rep. 2016. PMID: 27545962 Free PMC article.
-
The Multifaceted Role of Nutrient Sensing and mTORC1 Signaling in Physiology and Aging.Front Aging. 2021 Aug 27;2:707372. doi: 10.3389/fragi.2021.707372. eCollection 2021. Front Aging. 2021. PMID: 35822019 Free PMC article. Review.
-
Altered lysosomal positioning affects lysosomal functions in a cellular model of Huntington's disease.Eur J Neurosci. 2015 Aug;42(3):1941-51. doi: 10.1111/ejn.12957. Epub 2015 Jun 19. Eur J Neurosci. 2015. PMID: 25997742 Free PMC article.
-
Direct Interaction between Ras Homolog Enriched in Brain and FK506 Binding Protein 38 in Cashmere Goat Fetal Fibroblast Cells.Asian-Australas J Anim Sci. 2014 Dec;27(12):1671-7. doi: 10.5713/ajas.2014.14145. Asian-Australas J Anim Sci. 2014. PMID: 25358358 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

