Quantitative trait locus for body weight identified on rat chromosome 4 in inbred alcohol-preferring and -nonpreferring rats: potential implications for neuropeptide Y and corticotrophin releasing hormone 2
- PMID: 23312492
- PMCID: PMC3545280
- DOI: 10.1016/j.alcohol.2012.10.005
Quantitative trait locus for body weight identified on rat chromosome 4 in inbred alcohol-preferring and -nonpreferring rats: potential implications for neuropeptide Y and corticotrophin releasing hormone 2
Abstract
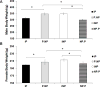
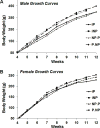
The alcohol-preferring (P) and -nonpreferring (NP) rat lines were developed using bidirectional selective breeding for alcohol consumption (g/kg/day) and alcohol preference (water:ethanol ratio). During a preliminary study, we detected a difference in body weight between inbred P (iP) and inbred NP (iNP) rats that appeared to be associated with the transfer of the Chromosome 4 quantitative trait locus (QTL) seen in the P.NP and NP.P congenic strains. After the initial confirmation that iP rats displayed lower body weight when compared to iNP rats (data not shown), body weight and growth rates of each chromosome 4 reciprocal congenic rat strain (P.NP and NP.P) were measured, and their body weight was consistent with their respective donor strain phenotype, confirming that a quantitative trait locus for body weight mapped to the chromosome 4 interval. Utilizing the newly developed interval-specific congenic strains (ISCS-A and ISCS-B), the QTL interval was further narrowed identifying the following candidate genes of interest: neuropeptide Y (Npy), juxtaposed with another zinc finger gene 1 (Jazf1), corticotrophin releasing factor receptor 2 (Crfr2) and LanC lantibiotic synthetase component C-like 2 (Lancl2). These findings indicate that a biologically active variant(s) regulates body weight on rat chromosome 4 in iP and iNP rats. This QTL for body weight was successfully captured in the P.NP and NP.P congenic strains, and interval-specific congenic strains (ISCSs) were subsequently employed to fine-map the QTL interval identifying the following candidate genes of interest: Npy, Jazf1, Crfr2 and Lancl2. Both Npy and Crfr2 have been previously identified as candidate genes of interest underlying the chromosome 4 QTL for alcohol consumption in iP and iNP rats.
Published by Elsevier Inc.
Figures

Similar articles
-
Development of congenic rat strains for alcohol consumption derived from the alcohol-preferring and nonpreferring rats.Behav Genet. 2006 Mar;36(2):285-90. doi: 10.1007/s10519-005-9021-z. Epub 2006 Feb 10. Behav Genet. 2006. PMID: 16470346
-
Candidate genes for alcohol preference identified by expression profiling in alcohol-preferring and -nonpreferring reciprocal congenic rats.Genome Biol. 2010;11(2):R11. doi: 10.1186/gb-2010-11-2-r11. Epub 2010 Feb 3. Genome Biol. 2010. PMID: 20128895 Free PMC article.
-
Alcohol-preferring rats show decreased corticotropin-releasing hormone-2 receptor expression and differences in HPA activation compared to alcohol-nonpreferring rats.Alcohol Clin Exp Res. 2014 May;38(5):1275-83. doi: 10.1111/acer.12379. Epub 2014 Mar 10. Alcohol Clin Exp Res. 2014. PMID: 24611993 Free PMC article.
-
From QTL to candidate gene: a genetic approach to alcoholism research.Curr Drug Abuse Rev. 2009 May;2(2):127-34. doi: 10.2174/1874473710902020127. Curr Drug Abuse Rev. 2009. PMID: 19630743 Free PMC article. Review.
-
A role for neuropeptide Y in alcohol intake control: evidence from human and animal research.Physiol Behav. 2003 Jun;79(1):95-101. doi: 10.1016/s0031-9384(03)00109-4. Physiol Behav. 2003. PMID: 12818714 Review.
Cited by
-
Npy deletion in an alcohol non-preferring rat model elicits differential effects on alcohol consumption and body weight.J Genet Genomics. 2016 Jul 20;43(7):421-30. doi: 10.1016/j.jgg.2016.04.010. Epub 2016 Apr 29. J Genet Genomics. 2016. PMID: 27461754 Free PMC article.
-
Estrogen-Dependent Upregulation of Adcyap1r1 Expression in Nucleus Accumbens Is Associated With Genetic Predisposition of Sex-Specific QTL for Alcohol Consumption on Rat Chromosome 4.Front Genet. 2018 Dec 4;9:513. doi: 10.3389/fgene.2018.00513. eCollection 2018. Front Genet. 2018. PMID: 30564267 Free PMC article.
References
-
- Alam I, Robling AG, Weissing S, Carr LG, Lumeng L, Turner CH. Bone mass and strength: phenotypic and genetic relationship to alcohol preference in P/NP and HAD/LAD rats. Alcohol Clin Exp Res. 2005;29:1769–1776. - PubMed
-
- Apter SJ, Eriksson CJ. The role of social isolation in the effects of alcohol on corticosterone and testosterone levels of alcohol-preferring and non-preferring rats. Alcohol Alcohol. 2006;41:33–38. - PubMed
-
- Bale TL, Vale WW. CRF and CRF receptors: role in stress responsivity and other behaviors. Annu Rev Pharmacol Toxicol. 2004;44:525–557. - PubMed
-
- Bice P, Foroud T, Bo R, Castelluccio P, Lumeng L, Li TK, Carr LG. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998;9:949–955. - PubMed
-
- Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg H, Lumeng L, Li TK. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22:884–887. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

