Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome
- PMID: 23313553
- PMCID: PMC3597350
- DOI: 10.1016/j.cell.2012.10.054
Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome
Erratum in
- Cell. 2015 Jun 18;161(7):1697-8
Abstract
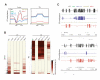
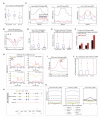
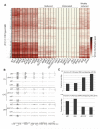
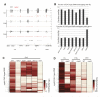
Cytosine methylation is involved in various biological processes such as silencing of transposable elements (TEs) and imprinting. Multiple pathways regulate DNA methylation in different sequence contexts, but the factors that regulate DNA methylation at a given site in the genome largely remain unknown. Here we have surveyed the methylomes of a comprehensive list of 86 Arabidopsis gene silencing mutants by generating single-nucleotide resolution maps of DNA methylation. We find that DNA methylation is site specifically regulated by different factors. Furthermore, we have identified additional regulators of DNA methylation. These data and analyses will serve as a comprehensive community resource for further understanding the control of DNA methylation patterning.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



References
-
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. - PMC - PubMed
-
- Baürle I, Smith L, Baulcombe DC, Dean C. Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science. 2007;318:109–112. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

