Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons
- PMID: 23313911
- PMCID: PMC3568452
- DOI: 10.1038/nn.3307
Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons
Abstract
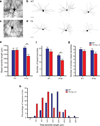
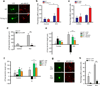
L-type voltage gated calcium channels have an important role in neuronal development by promoting dendritic growth and arborization. A point mutation in the gene encoding Ca(V)1.2 causes Timothy syndrome, a neurodevelopmental disorder associated with autism spectrum disorders (ASDs). We report that channels with the Timothy syndrome alteration cause activity-dependent dendrite retraction in rat and mouse neurons and in induced pluripotent stem cell (iPSC)-derived neurons from individuals with Timothy syndrome. Dendrite retraction was independent of calcium permeation through the mutant channel, was associated with ectopic activation of RhoA and was inhibited by overexpression of the channel-associated GTPase Gem. These results suggest that Ca(V)1.2 can activate RhoA signaling independently of Ca(2+) and provide insights into the cellular basis of Timothy syndrome and other ASDs.
Conflict of interest statement
The authors have no competing financial interests.
Figures



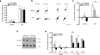

References
-
- McAllister AK, Katz LC, Lo DC. Neurotrophin regulation of cortical dendritic growth requires activity. Neuron. 1996;17:1057–1064. - PubMed
-
- Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. - PubMed
-
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. - PubMed
-
- Splawski I, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. - PubMed
-
- Dolmetsch R. Excitation-transcription coupling: signaling by ion channels to the nucleus. Sci STKE. 2003;2003:PE4. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

