Bioinformatics analysis of large-scale viral sequences: from construction of data sets to annotation of a phylogenetic tree
- PMID: 23314574
- PMCID: PMC3544756
- DOI: 10.4161/viru.23161
Bioinformatics analysis of large-scale viral sequences: from construction of data sets to annotation of a phylogenetic tree
Abstract
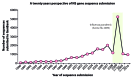
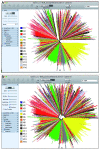
Due to a significant decrease in the cost of DNA sequencing, the number of sequences submitted to the public databases has dramatically increased in recent years. Efficient analysis of these data sets may lead to a significant understanding of the nature of pathogens such as bacteria, viruses, parasites, etc. However, this has raised questions about the efficacy of currently available algorithms for the study of pathogen evolution and construction of phylogenetic trees. While the advanced algorithms and corresponding programs are being developed, it is crucial to optimize the available ones in order to cope with the current need. The protocol presented in this study is optimized using a number of strategies currently being proposed for handling large-scale DNA sequence data sets, and offers a highly efficacious and accurate method for computing phylogenetic trees with limited computer resources. The protocol may take up to 36 h for construction and annotation of a final tree of about 20,000 sequences.
Figures



Similar articles
-
Building (Viral) Phylogenetic Trees Using a Maximum Likelihood Approach.Curr Protoc Microbiol. 2018 Nov;51(1):e63. doi: 10.1002/cpmc.63. Epub 2018 Sep 28. Curr Protoc Microbiol. 2018. PMID: 30265446
-
Computational framework for next-generation sequencing of heterogeneous viral populations using combinatorial pooling.Bioinformatics. 2015 Mar 1;31(5):682-90. doi: 10.1093/bioinformatics/btu726. Epub 2014 Oct 29. Bioinformatics. 2015. PMID: 25359889
-
Living Trees: High-Quality Reproducible and Reusable Construction of Bacterial Phylogenetic Trees.Mol Biol Evol. 2020 Feb 1;37(2):563-575. doi: 10.1093/molbev/msz241. Mol Biol Evol. 2020. PMID: 31633785
-
Bioinformatics Goes Viral: I. Databases, Phylogenetics and Phylodynamics Tools for Boosting Virus Research.Viruses. 2024 Sep 6;16(9):1425. doi: 10.3390/v16091425. Viruses. 2024. PMID: 39339901 Free PMC article. Review.
-
Origins and challenges of viral dark matter.Virus Res. 2017 Jul 15;239:136-142. doi: 10.1016/j.virusres.2017.02.002. Epub 2017 Feb 9. Virus Res. 2017. PMID: 28192164 Review.
Cited by
-
Estimation of evolutionary dynamics and selection pressure in coronaviruses.Methods Mol Biol. 2015;1282:41-8. doi: 10.1007/978-1-4939-2438-7_4. Methods Mol Biol. 2015. PMID: 25720469 Free PMC article.
References
-
- Swofford D, Olsen G, Waddel P, Hillis DM. Phylogenetic inference. Pages in (Molecular systematics, 2nd edition (D. M. Hillis, C.Moritz, and B. K. Mable, eds.). Sinauer, Sunderland, Massachusetts.
-
- Page R, Holmes E. Molecular evolution: A phylogenetic approach. Blackwell, Osney Mead, Oxford, UK.
-
- Jin L, Nei M. Limitations of the evolutionary parsimony method of phylogenetic analysis. Mol Biol Evol. 1990;7:82–102. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
